Genome Mining of α-Pyrone Natural Products from Ascidian-Derived Fungus Amphichordafelina SYSU-MS7908
Abstract
:1. Introduction
2. Results and Discussion
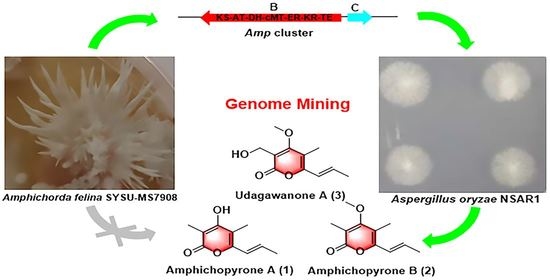
2.1. Bioinformatic Analysis of the Amp Cluster
2.2. Heterologous Expression of the Amp Cluster in A. oryzae
2.3. Characterization of amphichopyrone A (1) and B (2)
2.4. Evaluation of Anti-Inflammatory Activity
3. Materials and Methods
3.1. General Materials
3.2. Strains and Media
3.3. Construction of Recombinant Plasmids
3.4. Transformation of A. oryzae NSAR1
3.5. Extraction, Isolation, and Characterization
3.6. Anti-Inflammatory Activity
3.7. Quantification of the Expression of iNOS, COX-2, TNF-α, IL-6, and IL-1β and GAPDH
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodruff, H.B. Natural products from microorganisms. Science 1980, 208, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, J.; Mou, P.; Yan, Y.; Chen, M.; Tang, Y. Genome mining of cryptic tetronate natural products from a PKS-NRPS encoding gene cluster in Trichoderma harzianum t-22. Org. Biomol. Chem. 2021, 19, 1985–1990. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shu, S.; Kalaitzis, J.A.; Shang, Z.; Vuong, D.; Crombie, A.; Lacey, E.; Piggott, A.M.; Chooi, Y.H. Genome mining of Aspergillus hancockii unearths cryptic polyketide hancockinone A featuring a prenylated 6/6/6/5 carbocyclic skeleton. Org. Lett. 2021, 23, 8789–8793. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, X.; Sato, Y.; Zhu, G.; Minami, A.; Zhang, W.; Ozaki, T.; Zhu, B.; Wang, Z.; Wang, X.; et al. Genome-based discovery of enantiomeric pentacyclic sesterterpenes catalyzed by fungal bifunctional terpene synthases. Org. Lett. 2021, 23, 4645–4650. [Google Scholar] [CrossRef]
- Yan, D.; Matsuda, Y. Genome mining-driven discovery of 5-methylorsellinate-derived meroterpenoids from Aspergillus funiculosus. Org. Lett. 2021, 23, 3211–3215. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, M.; Chen, B.; Salaenoi, J.; Niaz, S.-I.; He, J.; Liu, L. Penicamide A, a unique N,N′-ketal quinazolinone alkaloid from ascidian-derived fungus Penicillium sp. 4829. Mar. Drugs 2019, 17, 522. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Shen, H.; Deng, Y.; Guo, H.; Jiang, M.; Wu, Z.; Yin, H.; Liu, L. Roussoelins A and B: Two phenols with antioxidant capacity from ascidian-derived fungus Roussoella siamensis SYSU-MS4723. Mar. Life Sci. Technol. 2021, 3, 69–76. [Google Scholar] [CrossRef]
- Chen, S.; Shen, H.; Zhang, P.; Cheng, H.; Dai, X.; Liu, L. Anti-glioma trichobamide A with an unprecedented tetrahydro-5H-furo[2,3-b]pyrrol-5-one functionality from ascidian-derived fungus Trichobotrys effuse 4729. Chem. Commun. 2019, 55, 1438–1441. [Google Scholar] [CrossRef]
- Niaz, S.-I.; Zhang, P.; Shen, H.; Li, J.; Chen, B.; Chen, S.; Liu, L.; He, J. Two new isochromane derivatives penisochromanes A and B from ascidian-derived fungus Penicillium sp. 4829. Nat. Prod. Res. 2019, 33, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.H.; Wu, Z.E.; Wu, Q.L.; Yin, H.M.; Guo, H.; Yuan, S.W.; Liu, Z.M.; Chen, S.H.; Liu, L. Amphichoterpenoids A-C, unprecedented picoline-derived meroterpenoids from the ascidian-derived fungus Amphichorda felina SYSU-MS7908. Chin. Chem. Lett. 2021, 32, 1893–1896. [Google Scholar] [CrossRef]
- Liang, M.; Lyu, H.N.; Ma, Z.Y.; Li, E.W.; Cai, L.; Yin, W.B. Genomics-driven discovery of a new cyclodepsipeptide from the guanophilic fungus Amphichorda guana. Org. Biomol. Chem. 2021, 19, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J. Recent Advances in the synthesis of 2-pyrones. Mar. Drugs 2015, 13, 1581–1620. [Google Scholar] [CrossRef] [Green Version]
- McGlacken, G.P.; Fairlamb, I.J. 2-Pyrone natural products and mimetics: Isolation, characterisation and biological activity. Nat. Prod. Rep. 2005, 22, 369–385. [Google Scholar] [CrossRef]
- Macabeo, A.P.G.; Cruz, A.J.C.; Narmani, A.; Arzanlou, M.; Babai-Ahari, A.; Pilapil, L.A.E.; Garcia, K.Y.M.; Huch, V.; Stadler, M. Tetrasubstituted α-pyrone derivatives from the endophytic fungus, Neurospora udagawae. Phytochem. Lett. 2020, 35, 147–151. [Google Scholar] [CrossRef]
- Kasahara, K.; Miyamoto, T.; Fujimoto, T.; Oguri, H.; Tokiwano, T.; Oikawa, H.; Ebizuka, Y.; Fujii, I. Solanapyrone synthase, a possible Diels-Alderase and iterative type I polyketide synthase encoded in a biosynthetic gene cluster from Alternaria solani. ChemBioChem 2010, 11, 1245–1252. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Zhong, Y.; Yu, Y.; Shi, D.F.; Huang, H.Y.; Tang, X.L.; Wang, Y.H.; Chen, G.D.; Zhang, H.P.; Liu, C.L.; et al. 4-Hydroxy pyridones from heterologous expression and cultivation of the native host. J. Nat. Prod. 2020, 83, 3338–3346. [Google Scholar] [CrossRef]
- Guo, J.; Cai, Y.S.; Cheng, F.; Yang, C.; Zhang, W.; Yu, W.; Yan, J.; Deng, Z.; Hong, K. Genome mining reveals a multiproduct sesterterpenoid biosynthetic gene cluster in Aspergillus ustus. Org. Lett. 2021, 23, 1525–1529. [Google Scholar] [CrossRef]
- Wang, W.-G.; Du, L.-Q.; Sheng, S.-L.; Li, A.; Li, Y.-P.; Cheng, G.-G.; Li, G.-P.; Sun, G.; Hu, Q.-F.; Matsuda, Y. Genome mining for fungal polyketide-diterpenoid hybrids: Discovery of key terpene cyclases and multifunctional P450s for structural diversification. Org. Chem. Front. 2019, 6, 571–578. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar]
- Kim, W.; Park, J.-J.; Gang, D.R.; Peever, T.L.; Chen, W. A novel type pathway-specific regulator and dynamic genome environments of a solanapyrone biosynthesis gene cluster in the fungus Ascochyta rabiei. Eukaryot. Eukaryot. Cell 2015, 14, 1102–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.; Park, C.-M.; Park, J.-J.; Akamatsu, H.O.; Peever, T.L.; Xian, M.; Gang, D.R.; Vandemark, G.; Chen, W. Functional analyses of the Diels-Alderase gene sol5 of Ascochyta rabiei and Alternaria solani indicate that the solanapyrone phytotoxins are not required for pathogenicity. Mol. Plant-Microbe Interact. 2015, 28, 482–496. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, H.; Satoh, Y.; Yamazaki, M. Four new immunosuppressive components, Kobiin and Kobifuranones A, B, and C, from an Ascomycete, Gelasinospora kobi. Chem. Pharm. Bull. 1998, 46, 211–216. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, H. Immunomodulatory constituents from Ascomycetous fungi. J. Nat. Med. 2018, 72, 20–31. [Google Scholar] [CrossRef]
- Sun, L.D.; Wang, F.; Dai, F.; Wang, Y.H.; Lin, D.; Zhou, B. Development and mechanism investigation of a new piperlongumine derivative as a potent anti-inflammatory agent. Biochem. Pharmacol. 2015, 95, 156–169. [Google Scholar] [CrossRef]
- Xue, G.M.; Li, X.Q.; Chen, C.; Chen, K.; Wang, X.B.; Gu, Y.C.; Luo, J.G.; Kong, L.Y. Highly oxidized guaianolide sesquiterpenoids with potential anti-inflammatory activity from Chrysanthemum indicum. J. Nat. Prod. 2018, 81, 378–386. [Google Scholar] [CrossRef]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef]
- Jin, F.J.; Maruyama, J.; Juvvadi, P.R.; Arioka, M.; Kitamoto, K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol. Lett. 2004, 239, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Fujii, T.; Yamaoka, H.; Gomi, K.; Kitamoto, K.; Kumaga, C. Cloning and nucleotide sequence of the ribonuclease T1 gene (rntA) from Aspergillus oryzae and its expression in Saccharomyces cerevisiae and Aspergillus oryzae. Biosci. Biotechnol. Biochem. 1995, 59, 1869–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubodera, T.; Yamashita, N.; Nishimura, A. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: Cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 2000, 64, 1416–1421. [Google Scholar] [CrossRef] [PubMed]
- Pall, M.L.; Brunelli, J.P. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newsl. 1993, 40, 59–62. [Google Scholar] [CrossRef] [Green Version]
- Jin, F.J.; Maruyama, J.-I.; Juvvadi, P.R.; Arioka, M.; Kitamoto, K. Adenine auxotrophic mutants of Aspergillus oryzae: Development of a novel transformation system with triple auxotrophic hosts. Biosci. Biotechnol. Biochem. 2004, 68, 656–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, O.; Lee, B.R.; Gomi, K.; Iimura, Y. Cloning and functional analysis of the Aspergillus oryzae conidiation regulator gene brlA by its disruption and misscheduled expression. J. Biosci. Bioeng. 1999, 87, 424–429. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Q.; Jin, X.; Zou, X.; Wang, Y.; Hao, D.; Fu, F.; Jiao, W.; Zhang, C.; Lin, H.; et al. Dysifragilone A inhibits LPS induced RAW264.7 macrophage activation by blocking the p38 MAPK signaling pathway. Mol. Med. Rep. 2018, 17, 674–682. [Google Scholar] [CrossRef] [Green Version]







| Amp | AA | Homolog (Accession No.) | S/I a (%) | Proposed Function |
|---|---|---|---|---|
| A | 285 | - | - | Methyltransferase |
| B | 2633 | Sol1 (D7UQ44.1) | 67.5/51.0 | polyketide synthase |
| C | 455 | Sol2 (XP_045265976.1) | 64.6/49.5 | O-methyltransferase |
| D | 583 | 1A4 (CRG92717.1) | 85.6/72.9 | p450 |
| E | 273 | DltE (XP_045265975.1) | 86.4/72.5 | Oxidoreductase |
| F | 518 | ChyH (XP_037172406.1) | 80.6/67.4 | FAD-linked oxidoreductase |
| G | 487 | — | — | p450 |
| H | 422 | — | — | Unknown |
| I | 398 | — | — | Unknown |
| J | 253 | CsgA (CZT51343.1) | 70.8/52.2 | short-chain dehydrogenase/reductase |
| No | 1 | 2 | ||
|---|---|---|---|---|
| δH,(J in Hz) | δC, Type | δH,(J in Hz) | δC, Type | |
| 2 | 163.6, C | 164.5, C | ||
| 3 | 99.0, C | 111.2, C | ||
| 4 | 164.3, C | 168.7, C | ||
| 5 | 106.3, C | 109.4, C | ||
| 6 | 151.6, C | 153.0, C | ||
| 7 | 6.42, dq (15.4, 1.3) | 120.6, CH | 6.42, dq (15.4, 1.3) | 121.4, CH |
| 8 | 6.50, dq (15.4, 6.0) | 132.0, CH | 6.51, dq (15.4, 6.5) | 133.1, CH |
| 9 | 1.90, d (6.0) | 17.6, CH3 | 1.91, d (6.5) | 18.6, CH3 |
| 10 | 1.94, s | 8.6, CH3 | 1.96, s | 10.4, CH3 |
| 11 | 2.01, s | 8.4, CH3 | 1.98, s | 9.5, CH3 |
| 12 | 3.83, s | 60.7, CH3 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, S.; Chen, L.; Wu, Q.; Jiang, M.; Guo, H.; Hu, Z.; Chen, S.; Liu, L.; Gao, Z. Genome Mining of α-Pyrone Natural Products from Ascidian-Derived Fungus Amphichordafelina SYSU-MS7908. Mar. Drugs 2022, 20, 294. https://doi.org/10.3390/md20050294
Yuan S, Chen L, Wu Q, Jiang M, Guo H, Hu Z, Chen S, Liu L, Gao Z. Genome Mining of α-Pyrone Natural Products from Ascidian-Derived Fungus Amphichordafelina SYSU-MS7908. Marine Drugs. 2022; 20(5):294. https://doi.org/10.3390/md20050294
Chicago/Turabian StyleYuan, Siwen, Litong Chen, Qilin Wu, Minghua Jiang, Heng Guo, Zhibo Hu, Senhua Chen, Lan Liu, and Zhizeng Gao. 2022. "Genome Mining of α-Pyrone Natural Products from Ascidian-Derived Fungus Amphichordafelina SYSU-MS7908" Marine Drugs 20, no. 5: 294. https://doi.org/10.3390/md20050294
APA StyleYuan, S., Chen, L., Wu, Q., Jiang, M., Guo, H., Hu, Z., Chen, S., Liu, L., & Gao, Z. (2022). Genome Mining of α-Pyrone Natural Products from Ascidian-Derived Fungus Amphichordafelina SYSU-MS7908. Marine Drugs, 20(5), 294. https://doi.org/10.3390/md20050294