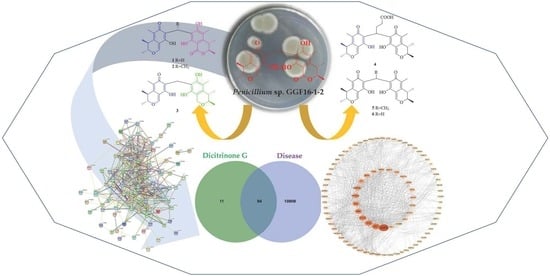
Rare Carbon-Bridged Citrinin Dimers from the Starfish-Derived Symbiotic Fungus Penicillium sp. GGF16-1-2
Abstract
:1. Introduction
2. Results
2.1. Structural Identification of New Compounds
2.2. Evaluation of Antifungal Activity
2.3. Cytotoxic Assays
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Materials, Extraction, and Fermentation
3.3. Isolation
3.4. Structural Characterizations of the New Compounds 1–4
3.5. Antifungal Activity Assay
3.6. Cytotoxic Assays
3.7. Quantum Chemical Calculations
3.8. Targets Prediction
3.9. Western Blot Assays
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, Y.Q.; Shen, J.J.; Liu, Z.; Xia, K.Y.; Zhu, W.M.; Fu, P. Methylene-bridged dimeric natural products involving one-carbon unit in biosynthesis. Nat. Prod. Rep. 2022, 39, 1305–1324. [Google Scholar] [CrossRef] [PubMed]
- Wezeman, T.; Brase, S.; Masters, K.S. Xanthone dimers: A compound family which is both common and privileged. Nat. Prod. Rep. 2015, 32, 6–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.X.; Dias, J.R. Dimeric and Oligomeric Steroids. Chem. Rev. 1997, 97, 283–304. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, C.L.; Yu, G.H.; Sun, Z.C.; Zhang, G.J.; Gu, Q.Q.; Zhu, T.J.; Che, C.; Guan, H.S.; Li, D.H. Dicitrinones E and F, citrinin dimers from the marine derived fungus Penicillium citrinum HDN-152-088. Tetrahedron Lett. 2019, 60, 151182. [Google Scholar] [CrossRef]
- Du, L.; Liu, H.C.; Fu, W.; Li, D.H.; Pan, Q.M.; Zhu, T.J.; Geng, M.Y.; Gu, Q.Q. Unprecedented citrinin trimer tricitinol B functions as a novel topoisomerase IIα inhibitor. J. Med. Chem. 2011, 54, 5796–5810. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Sadhasivam, S.; Barda, O.; Zakin, V.; Reifen, R.; Sionov, E. Rapid detection and quantification of patulin and citrinin contamination in fruits. Molecules 2021, 26, 4545. [Google Scholar] [CrossRef]
- Wang, W.Y.; Liao, Y.Y.; Zhang, B.B.; Gao, M.L.; Ke, W.Q.; Li, F.; Shao, Z.Z. Citrinin monomer and dimer derivatives with antibacterial and cytotoxic activities isolated from the deep sea-derived fungus Penicillium citrinum NLG-S01-P1. Mar. Drugs 2019, 17, 46. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.; Sun, T.T.; Yang, J.K.; Zhao, G.Z.; Liu, Q.A.; Hu, L.D.; Ma, Z.Y.; Zhu, H.J. The absolute configuration of anti-vibrio citrinin dimeric derivative by VCD, ECD and NMR methods. Nat. Prod. Res. 2019, 33, 2192–2199. [Google Scholar] [CrossRef]
- Ni, M.; Lin, W.L.; Yang, P.; Mao, S.C. A novel citrinin derivative from the marine-source fungus Penicillium citrinum. Acta Pharm. Sin. 2015, 50, 203–206. [Google Scholar]
- Wei, X.; Su, J.C.; Hu, H.S.; He, X.X.; Lin, S.J.; Zhang, D.M.; Ye, W.C.; Chen, M.F.; Lin, H.W.; Zhang, C.X. Probing indole diketopiperazine−based hybrids as environmental-2 induced products from Aspergillus sp. EGF 15-0-3. Org. Lett. 2022, 24, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Su, J.C.; Liu, Y.H.; Deng, B.; Hu, Z.F.; Wu, J.L.; Xia, R.F.; Chen, C.; He, Q.; Chen, J.C.; et al. Stelleranoids A–M, guaiane-type sesquiterpenoids based on [5,7] bicyclic system from Stellera chamaejasme and their cytotoxic activity. Bioorg. Chem. 2021, 115, 105251. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, F.T.; Si-Tu, M.X.; Fan, H.; Hu, J.S.; Yang, H.; Guan, S.Y.; An, L.K.; Zhang, C.X. Pyranodipyran derivatives with tyrosyl DNA phosphodiesterase 1 inhibitory activities and fluorescent properties from Aspergillus sp. EGF 15-0-3. Mar. Drugs 2022, 20, 211. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, D.H.; Zhang, G.J.; Zhu, T.J.; Ai, J.; Gu, Q.Q. Novel carbon-bridged citrinin dimers from a volcano ash-derived fungus Penicillium citrinum and their cytotoxic and cell cycle arrest activities. Tetrahedron 2010, 66, 9286–9290. [Google Scholar] [CrossRef]
- Trivedi, A.B.; Hirota, M.; Doi, E.; Kitabatake, N.F. Formation of a new toxic compound, citrinin Hl, from citrinin on mild heating in water. J. Chem. Soc. Perkin Trans. 1 1993, 24, 2167–2171. [Google Scholar] [CrossRef]
- Filho, J.W.G.O.; Islam, M.T.; Ali, E.S.; Uddin, S.J.; Santos, J.V.O.; Marcus, V.B.O.A.; Junior, A.L.G.; Marcia, F.C.J.P.; Maria, D.R.M.B.; Joao, M.E.S.C.; et al. A comprehensive review on biological properties of citrinin. Food Chem. Toxicol. 2017, 110, 130–141. [Google Scholar] [CrossRef]
- Liu, H.C.; Du, L.; Zhu, T.J.; Li, D.H.; Geng, M.Y.; Gu, Q.Q. Two new citrinin dimers from a volcano ash-derived fungus, Penicillium citrinum HGY1-5. Helv. Chim. Acta 2010, 93, 2224–2230. [Google Scholar] [CrossRef]
- Benjamin, R.C.; Robert, J.C.; Ernest, L.; Shaun, T.; Jennifer, H.G. Citrinin revisited: From monomers to dimers and beyond. Org. Biom. Chem. 2006, 4, 1520–1528. [Google Scholar]
- Wakana, D.; Hosoe, T.; Itabashi, T.; Okada, K.; Takaki, C.; Galba, M.; Yaguchi, T.; Fukushima, K.; Kawai, K. New citrinin derivatives isolated from Penicillium citrinum. J. Nat. Med. 2006, 60, 279–284. [Google Scholar] [CrossRef]
- Wu, C.J.; Yi, L.; Cui, C.B.; Cui, C.B.; Li, C.W.; Wang, N.; Han, X. Activation of the silent secondary metabolite production by introducing neomycin-resistance in a marine-derived Penicillium purpurogenum G59. Mar. Drugs 2015, 13, 2465–2487. [Google Scholar] [CrossRef] [Green Version]
- Hill, R.K.; Gardella, L.A. The absolute configuration of citrinin. J. Org. Chem. 1964, 29, 128–134. [Google Scholar] [CrossRef]
- Hu, Y.M.; Zhang, J.N.; Liu, D.; Guo, D.; Liu, T.X.; Xin, Z.H. Pencitrin and pencitrinol, two new citrinin derivatives from an endophytic fungus Penicillium citrinum salicorn 46. Phytochem. Lett. 2017, 22, 229–234. [Google Scholar] [CrossRef]
- Xin, Z.H.; Tian, L.; Zhu, T.J.; Wang, W.L.; Du, L.; Fang, Y.C.; Gu, Q.Q.; Zhu, W.M. Isocoumarin derivatives from the sea squirt-derived fungus penicillium stoloniferum QY2-10 and the halotolerant fungus penicillium notatum B-52. Arch. Pharm. Res. 2007, 30, 816–819. [Google Scholar] [CrossRef]
- Shen, B. Biosynthesis of aromatic polyketides. In Biosynthesis; Topics in Current Chemistry; Springer: Berlin/Heidelberg, Germany, 2000; Volume 209, pp. 1–51. [Google Scholar]
- Friedel, C.; Crafts, J.M. The alkylation or acylation of aromatic compounds catalyzed by aluminum chloride or other Lewis acids. Compt. Rend. 1877, 84, 1392. [Google Scholar]
- Wang, T.; Ren, D.D.; Guo, H.; Chen, X.; Zhu, P.K.; Nie, H.Z.; Xu, L. CgSCD1 is essential for melanin biosynthesis and pathogenicity of Colletotrichum gloeosporioides. Pathogens 2020, 20, 141. [Google Scholar] [CrossRef] [Green Version]
- Shao, X.H.; Lai, D.; Zhu, H.X.; Huang, J.H.; Li, S.M.; Kuang, S.Z. Virulence and control efficacy infield of different fungicides against the pathogen of guava shoot blight. Plant Protect. 2019, 45, 199–203, 217. [Google Scholar]
- Dorman, K.; Heinemann, V.; Kobold, S.; Bergwelt-Baildon, M. Novel systemic treatment approaches for metastatic pancreatic cancer. Expert Opin. Investig. Drugs 2022, 31, 249–262. [Google Scholar] [CrossRef]
- Tim, M. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2019, 47, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Tao, H.; Jin, C.; Du, Z.Y.; Liao, W.F.; Tang, Q.J.; Ding, K. Cordycepin inhibits pancreatic cancer cell growth in vitro and in vivo via targeting FGFR2 and blocking ERK signaling. Chin. J. Med. 2020, 18, 345–355. [Google Scholar] [CrossRef]
- Fujita, M.; Hasegawa, A.; Yamamori, M.; Okamura, N. In vitro and in vivo cytotoxicity of troglitazone in pancreatic cancer. J. Exp. Clin. Cancer Res. 2017, 36, 91. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.X.; Zhang, L.; Thu, P.M.; Min, W.J.; Yang, P.; Li, J.; Li, P.; Xu, X.J. Sodium cantharidinate, a novel anti-pancreatic cancer agent that activates functional p53. Sci. China Life Sci. 2020, 64, 1295–1310. [Google Scholar] [CrossRef]
- Wei, X.; Feng, C.; Li, X.H.; Mao, X.X.; Luo, H.B.; Zhang, D.M.; Rong, L.; Xie, Z.Y.; Yu, X.; Li, J.; et al. Enantiomeric polyketides from the starfish-derived Symbiotic Fungus Penicillium sp. GGF16-1-2. Chem. Biodivers. 2019, 16, e1900052. [Google Scholar] [CrossRef]
- Li, X.D.; Su, J.C.; Jiang, B.Z.; Li, Y.L.; Guo, Y.Q.; Zhang, P. Janthinoid A, an unprecedented tri-nor-meroterpenoid with highly modified bridged 4a,1-(epoxymethano) phenanthrene scaffold, produced by the endophyte of Penicillium janthinellum TE-43. Org. Chem. Front. 2021, 8, 6196–6202. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. (Eds.) Gaussian 09, Revision A.02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Song, J.G.; Su, J.C.; Song, Q.Y.; Huang, R.L.; Tang, W.; Hu, L.J.; Huang, X.J.; Jiang, R.W.; Li, Y.L.; Ye, W.C.; et al. Cleistocaltones A and B, antiviral phloroglucinol-terpenoid adducts from Cleistocalyx operculatus. Org. Lett. 2019, 21, 9579–9583. [Google Scholar] [CrossRef]
- Su, J.C.; Wang, S.; Cheng, W.; Huang, X.J.; Li, M.M.; Jiang, E.W.; Li, Y.L.; Wang, L.; Ye, W.C.; Wang, Y. Phloroglucinol derivatives with unusual skeletons from Cleistocalyx operculatus and their in vitro antiviral activity. J. Org. Chem. 2018, 83, 8522–8532. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumloffel, A.; Hemberger, Y.; Pescitelli, G. SpecDis Version 1.70; University of Wuerzburg: Wuerzburg, Germany, 2017. [Google Scholar]





| No. | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δH, Mult, J | δC, Mult | δH, Mult, J | δC, Mult | δH, Mult, J | δC, Mult | δH, Mult, J | δC, Mult | |
| 1 | 8.09, s | 157.7, CH | 8.07, s | 158.7, CH | 8.00, s | 157.7, CH | 8.00, s/8.01, s | 158.0/158.2, CH |
| 3 | 4.74, dq (6.6, 13.4) | 80.1, CH | 4.69, dq (6.4, 12.9) | 79.8, CH | 4.70, dq (6.6, 13.1) | 79.9, CH | 4.72, m/4.72, m | 79.8/80.0, CH |
| 4 | 3.03, dq (7.2, 13.4) | 33.6, CH | 2.99, dq (7.1, 12.9) | 33.5, CH | 2.99, dq (7.2, 13.1) | 33.5, CH | 2.99, m/2.99, m | 33.5, CH |
| 4a | 135.5, C | 136.2, C | 136.0, C | 136.2/136.3, C | ||||
| 5 | 126.6, C | 126.6, C | 124.4, C | 124.2/124.6, C | ||||
| 6 | 187.3, C | 187.8, C | 184.5, C | 186.1/186.3, C | ||||
| 7 | 113.0, C | 117.1, C | 112.8, C | 116.1/116.2, C | ||||
| 8 | 161.9, C | 160.6, C | 161.5, C | 163.4/164.0, C | ||||
| 8a | 107.3, C | 107.0, C | 106.9, C | 106.8/106.9, C | ||||
| 9 | 1.17, d (6.6) | 19.5, CH3 | 1.22, d (6.4) | 19.4, CH3 | 1.18, d (6.6) | 17.6, CH3 | 1.22, d (6.6)/1.22, d (6.6) | 17.5/17.5, CH3 |
| 10 | 1.20, d (7.2) | 17.6, CH3 | 1.16, d (7.1) | 17.6 CH3 | 1.06, d (7.2) | 18.3, CH3 | 1.11, d (7.1)/1.11, d (7.1) | 18.2/18.3, CH3 |
| 11 | 1.90, s | 10.0, CH3 | 1.88, s | 9.9, CH3 | 1.89, s | 10.0, CH3 | 1.87, s/1.87, s | 9.7/9.8, CH3 |
| 1′ | 168.8, C | 168.9, C | 4.49, s | 59.0, CH2 | 8.03, s/8.04, s | 158.8/159.0, CH | ||
| 3′ | 4.71, dq (6.6, 13.4) | 80.1, CH | 4.72, dq (6.5, 12.9) | 79.8, CH | 3.77, m | 73.3, CH | 4.73, m/4.73, m | 80.1/80.1, CH |
| 4′ | 3.08, dq (7.2, 13.4) | 33.6, CH | 3.05, dq (7.1, 12.9) | 33.5, CH | 2.50, m | 34.5, CH | 2.99, m/2.99, m | 33.5, CH |
| 4a′ | 140.7, C | 140.2, C | 135.1, C | 136.9/136.9, C | ||||
| 5′ | 115.7, C | 117.1, C | 114.1, C | 125.0/125.3, C | ||||
| 6′ | 161.9, C | 162.7, C | 150.5, C | 186.6/186.6, C | ||||
| 7′ | 111.7, C | 115.7, C | 113.8, C | 116.3/116.6, C | ||||
| 8′ | 157.7, C | 158.7, C | 147.9, C | 163.8/163.8, C | ||||
| 8a′ | 98.0, C | 97.6, C | 114.6, C | 107.2/107.4, C | ||||
| 9′ | 1.07, d (6.6) | 18.4, CH3 | 1.09, d (6.5) | 18.4, CH3 | 1.08, d (6.6) | 17.8, CH3 | 1.15, d (6.6)/1.15, d (6.6) | 17.7/17.7, CH3 |
| 10′ | 1.21, d (7.2) | 19.6, CH3 | 1.17, d (7.1) | 19.6, CH3 | 1.09, d (7.2) | 20.5, CH3 | 1.04, d (7.1)/1.04, d (7.1) | 18.5/18.5, CH3 |
| 11′ | 2.04, s | 10.3, CH3 | 2.02, s | 10.4, CH3 | 2.02, s | 11.0, CH3 | 1.87, s/1.87, s | 10.1/10.1, CH3 |
| 1″ | a 3.61, d (8.6) b 3.67, d (8.6) | 17.2, CH2 | 4.89, q (7.5) | 24.1, CH | 3.57, br s | 18.1, CH2 | 4.29, t (7.2)/4.32, t (7.2) | 30.5/30.4, CH |
| 2″ | 1.55, d (7.4) | 16.4, CH3 | 2.36, m/2.36, m | 23.6/23.9, CH2 | ||||
| 3″ | 2.04, m/2.04, m | 32.7, CH2 | ||||||
| 4″ | 174.0/174.1, C | |||||||
| 8-OH | 13.15, s | |||||||
| 6′-OH | 12.39, s | 12.90, s | ||||||
| 8′-OH | 13.33, s | |||||||
| Compd. | Colletotrichum gloeosporioides | Compd. | Colletotrichum gloeosporioides |
|---|---|---|---|
| 1 | 16.14 | 6 | 0.61 |
| 2 | 10.23 | 7 | 5.31 |
| 3 | 9.58 | 8 | 7.58 |
| 4 | 9.63 | 9 | 4.34 |
| 5 | 8.87 | Carbendazim * | 49.58 |
| Compd. | BXPC-3 | PANC-1 |
|---|---|---|
| 1 | 12.25 ± 2.85 | 24.33 ± 2.10 |
| 2 | >50 | 39.54 ± 2.50 |
| 3 | >50 | >50 |
| 4 | >50 | >50 |
| 5 | >50 | >50 |
| 6 | >50 | >50 |
| 7 | 32.25 ± 3.82 | 49.85 ± 1.11 |
| 8 | >50 | >50 |
| 9 | >50 | >50 |
| Doxorubicin hydrochloride * | 18.24 ± 2.84 | 24.00 ± 3.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, H.; Shi, Z.-M.; Lei, Y.-H.; Si-Tu, M.-X.; Zhou, F.-G.; Feng, C.; Wei, X.; Shao, X.-H.; Chen, Y.; Zhang, C.-X. Rare Carbon-Bridged Citrinin Dimers from the Starfish-Derived Symbiotic Fungus Penicillium sp. GGF16-1-2. Mar. Drugs 2022, 20, 443. https://doi.org/10.3390/md20070443
Fan H, Shi Z-M, Lei Y-H, Si-Tu M-X, Zhou F-G, Feng C, Wei X, Shao X-H, Chen Y, Zhang C-X. Rare Carbon-Bridged Citrinin Dimers from the Starfish-Derived Symbiotic Fungus Penicillium sp. GGF16-1-2. Marine Drugs. 2022; 20(7):443. https://doi.org/10.3390/md20070443
Chicago/Turabian StyleFan, Hao, Zhi-Mian Shi, Yan-Hu Lei, Mei-Xia Si-Tu, Feng-Guo Zhou, Chan Feng, Xia Wei, Xue-Hua Shao, Yang Chen, and Cui-Xian Zhang. 2022. "Rare Carbon-Bridged Citrinin Dimers from the Starfish-Derived Symbiotic Fungus Penicillium sp. GGF16-1-2" Marine Drugs 20, no. 7: 443. https://doi.org/10.3390/md20070443
APA StyleFan, H., Shi, Z. -M., Lei, Y. -H., Si-Tu, M. -X., Zhou, F. -G., Feng, C., Wei, X., Shao, X. -H., Chen, Y., & Zhang, C. -X. (2022). Rare Carbon-Bridged Citrinin Dimers from the Starfish-Derived Symbiotic Fungus Penicillium sp. GGF16-1-2. Marine Drugs, 20(7), 443. https://doi.org/10.3390/md20070443