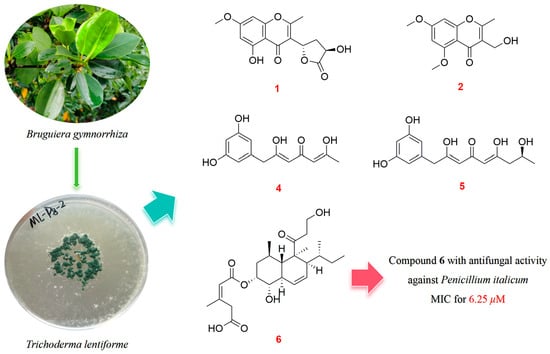
The Polyketides with Antimicrobial Activities from a Mangrove Endophytic Fungus Trichoderma lentiforme ML-P8-2
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Identification
2.2. Antimicrobial Assays
2.3. AChE Inhibitory Activity Assays
2.4. Cytotoxic Assays
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction, and Isolation
3.4. Alkali-hydrolysis Treatments for Compounds 6 and 8
3.5. ECD and Optical Rotation Computation Methods
3.6. Antimicrobial Assays
3.7. AChE Inhibition Assays
3.8. Cytotoxic Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, J. Bioactive natural products derived from mangrove-associated microbes. RSC Adv. 2015, 5, 841–892. [Google Scholar] [CrossRef]
- Chen, S.H.; Cai, R.L.; Liu, Z.M.; Cui, H.; She, Z.G. Secondary metabolites from mangrove-associated fungi: Source, chemistry and bioactivities. Nat. Prod. Rep. 2022, 39, 560–595. [Google Scholar] [CrossRef]
- Guo, R.; Li, G.; Zhang, Z.; Peng, X. Structures and biological activities of secondary metabolites from Trichoderma harzianum. Mar. Drugs 2022, 20, 701. [Google Scholar] [CrossRef]
- Guo, Q.; Shi, L.; Wang, X.; Li, D.; Yin, Z.; Zhang, J.; Ding, G.; Chen, L. Structures and biological activities of secondary metabolites from the Trichoderma genus (covering 2018–2022). J. Agric. Food Chem. 2023, 71, 13612–13632. [Google Scholar] [CrossRef] [PubMed]
- Su, D.Q.; Ding, L.J.; He, S. Marine-derived Trichoderma species as a promising source of bioactive secondary metabolites. Mini-Rev. Med. Chem. 2018, 18, 1702–1713. [Google Scholar] [CrossRef]
- Lai, C.; Chen, J.; Liu, J.; Tian, D.; Lan, D.; Liu, T.; Wu, B.; Bi, H.; Tang, J. New polyketides from a hydrothermal vent sediment fungus Trichoderma sp. JWM29-10-1 and their antimicrobial effects. Mar. Drugs 2022, 20, 720. [Google Scholar] [CrossRef]
- Yamada, T.; Mizutani, Y.; Umebayashi, Y.; Inno, N.; Kawashima, M.; Kikuchi, T.; Tanaka, R. Tandyukisin, a novel ketoaldehyde decalin derivative, produced by a marine sponge-derived Trichoderrna harzianum. Tetrahedron Lett. 2014, 55, 662–664. [Google Scholar] [CrossRef]
- Yamada, T.; Umebayashi, Y.; Kawashima, M.; Sugiura, Y.; Kikuchi, T.; Tanaka, R. Determination of the chemical structures of tandyukisins B-D, isolated from a marine sponge-derived fungus. Mar. Drugs 2015, 13, 3231–3240. [Google Scholar] [CrossRef] [PubMed]
- Suzue, M.; Kikuchi, T.; Tanaka, R.; Yamada, T. Tandyukisins E and F, novel cytotoxic decalin derivatives isolated from a marine sponge-derived fungus. Tetrahedron Lett. 2016, 57, 5070–5073. [Google Scholar] [CrossRef]
- Robertsen, H.L.; Musiol-Kroll, E.M. Actinomycete-derived polyketides as a source of antibiotics and lead structures for the development of new antimicrobial drugs. Antibiotics 2019, 8, 157. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yang, W.; Chen, T.; Tan, Q.; Zou, G.; Zang, Z.; Li, J.; Wang, B.; She, Z. Cytosporones W and X: Two mutually converting epimers from a mangrove endophytic fungus Diaporthe sp. ZJHJYZ-1. ACS Omega 2023, 8, 26628–26634. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Huang, H.; Shao, C.; Xia, X.; Ma, L.; Huang, X.; Lu, Y.; Lin, Y.; Long, Y.; She, Z. Asperterpenols A and B, new sesterterpenoids isolated from a mangrove endophytic fungus Aspergillus sp. 085242. Org. Lett. 2013, 15, 2522–2525. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.; Huang, Y.; Liu, L.; He, J.; Wang, L.; Yuan, J.; She, Z. Ascomylactams A–C, cytotoxic 12- or 13-membered-ring macrocyclic alkaloids isolated from the mangrove endophytic fungus Didymella sp. CYSK-4, and structure revisions of Phomapyrrolidones A and C. J. Nat. Prod. 2019, 82, 1752–1758. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, Y.; Tanahashi, T.; Nagakura, N.; Hamada, N. 2,3-Dialkylchromones from mycobiont cultures of the lichen Graphis scripta. Heterocycles 2000, 53, 1589–1593. [Google Scholar]
- Kobayashi, M.; Uehara, H.; Matsunami, K.; Aoki, S.; Kitagawa, I. Trichoharzin, a new polyketide produced by the imperfect fungus Trichoderma harzianum separated from the marine sponge Micale cecilia. Tetrahedron Lett. 1993, 34, 7925–7928. [Google Scholar] [CrossRef]
- Nakadate, S.; Nozawa, K.; Horie, H.; Fujii, Y.; Nagai, M.; Hosoe, T.; Kawai, K.I.; Yaguchi, T.; Fukushima, K. Eujavanicols A-C, decalin derivatives from Eupenicillium javanicum. J. Nat. Prod. 2007, 70, 1510–1512. [Google Scholar] [CrossRef] [PubMed]
- Duh, C.Y.; Wang, S.K.; Chung, S.G.; Chou, G.C.; Dai, C.F. Cytotoxic cembrenolides and steroids from the formosan soft coral Sarcophyton crassocaule. J. Nat. Prod. 2000, 63, 1634–1637. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Pierce, C.G.; Uppuluri, P.; Tristan, A.R.; Wormley, F.L., Jr.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef]




| Position | 6 | Position | 6 | ||
|---|---|---|---|---|---|
| δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | ||
| 1 | 58.1, CH2 | 3.85, ddd (11.0, 6.0, 3.8) 3.93, ddd (11.0, 7.2, 3.4) | 14 | 37.3, CH | 1.14, m |
| 2 | 41.3, CH2 | 2.69, ddd (18.9, 6.0, 3.4) 2.88, ddd (18.9, 7.2, 3.8) | 15 | 24.6, CH2 | 0.75, overlap 1.48, d (7.0) |
| 3 | 215.7, C | 16 | 12.7, CH3 | 0.77, d (4.2) | |
| 4 | 52.6, C | 17 | 19.4, CH3 | 0.95, d (6.7) | |
| 5 | 43.1, CH | 1.97, d (4.1) | 18 | 22.4, CH3 | 0.60, d (5.8) |
| 6 | 31.6, CH | 1.62, overlap | 19 | 19.5, CH3 | 1.28, s |
| 7 | 39.1, CH2 | 1.59, overlap 1.89, dd (12.1, 2.8) | 1’ | 167.5, C | |
| 8 | 73.6, CH | 5.30, m | 2’ | 119.6, CH | 6.01, s |
| 9 | 74.4, CH | 3.58, dd (10.9, 3.2) | 3’ | 151.9, C | |
| 10 | 40.4, CH | 2.12, d (10.9) | 4’ | 39.8, CH2 | 3.63, d (14.6) 3.77, d (14.6) |
| 11 | 125.7, CH | 6.05, d (10.6) | 5’ | 172.8, C | |
| 12 | 124.1, CH | 5.72, ddd (10.6, 4.7, 2.6) | 6’ | 26.2, CH3 | 2.06, s |
| 13 | 52.5, CH | 1.97, d (4.7) | |||
| MIC of Compounds/μM | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Euj. A 1 | Amp. 2 | Ket. 3 | |
| MRSA | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 0.25 | NT 4 |
| S. aureus | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 50 | >100 | >100 | 0.25 | NT |
| B. subtilis | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 0.25 | NT |
| S. typhimurium | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 50 | >100 | >100 | >100 | 0.25 | NT |
| P. aeruginosa | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 50 | 50 | >100 | >100 | 0.13 | NT |
| C. albicans | 50 | 100 | 25 | >100 | >100 | 25 | 50 | 50 | 25 | 25 | 25 | NT | 0.13 |
| P. italicum | >100 | >100 | >100 | >100 | >100 | 6.25 | 12.5 | 12.5 | 6.25 | 50 | 25 | NT | 1.56 |
| Compounds | IC50/μM | Compounds | IC50/μM |
|---|---|---|---|
| 1 | 38.6 ± 0.2 | 7 | 38.3 ± 0.4 |
| 2 | 33.7 ± 0.4 | 8 | 77.9 ± 1.7 |
| 3 | 20.6 ± 0.3 | 9 | 43.6 ± 0.4 |
| 4 | 37.7 ± 0.6 | 10 | 50.9 ± 0.5 |
| 5 | 51.3 ± 0.5 | Eujavanicol A | 32.4 ± 0.7 |
| 6 | 40.2 ± 0.7 | Donepezil Hydrochloride 1 | 65.5 ± 1.5 (nM) |
| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | |
| 2 | 168.0, C | 165.5, C | ||
| 3 | 117.8, C | 121.6, C | ||
| 4 | 182.1, C | 178.2, C | ||
| 4a | 105.1, C | 109.0, C | ||
| 5 | 163.2, C | 162.2, C | ||
| 6 | 99.3, CH | 6.32, d (2.2) | 97.1, CH | 6.50, d (2.3) |
| 7 | 167.4, C | 166.1, C | ||
| 8 | 93.3, CH | 6.49, d (2.2) | 93.9, CH | 6.59, d (2.3) |
| 8a | 159.0, C | 161.1, C | ||
| 9 | 56.5, CH3 | 3.86, s | 56.4, CH3 | 3.91, s |
| 10 | 18.1, CH3 | 2.52, s | 17.8, CH3 | 2.48, s |
| 11 | 55.5, CH2 | 4.57, s | ||
| 12 | 56.5, CH3 | 3.90, s | ||
| 2′ | 180.2, C | |||
| 3′ | 69.5, CH | 4.90, dd (9.3, 5.9) | ||
| 4′ | 36.2, CH2 | 2.48, m 2.75, ddd (13.5, 9.3, 4.1) | ||
| 5′ | 74.9, CH | 5.73, dd (9.9, 4.1) | ||
| Position | 4 | 5 | ||
|---|---|---|---|---|
| δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | |
| 1 | 106.9, CH | 6.12, d (1.9) | 106.9, CH | 6.12, d (2.1) |
| 2 | 158.5, C | 158.5, C | ||
| 3 | 101.2, CH | 6.10, d (1.9) | 101.2, CH | 6.10, d (2.1) |
| 4 | 158.5, C | 158.5, C | ||
| 5 | 106.9, CH | 6.12, d (1.9) | 106.9, CH | 6.12, d (2.1) |
| 6 | 137.5, C | 137.5, C | ||
| 7 | 38.7, CH2 | 3.65, s | 38.8, CH2 | 3.64, s |
| 8 | 167.6, C | 167.7, C | ||
| 9 | 113.2, CH | 6.04, d (2.3) | 113.3, CH | 6.04, d (2.3) |
| 10 | 178.7, C | 178.7, C | ||
| 11 | 113.2, CH | 6.07, dd (2.3, 0.9) | 114.0, CH | 6.05, d (2.3) |
| 12 | 165.8, C | 166.9, C | ||
| 13 | 19.2, CH3 | 2.20, d (0.9) | 42.6, CH2 | 2.52, d (3.7) |
| 14 | 64.0, CH | 3.90, m | ||
| 15 | 23.2, CH3 | 1.06, d (6.1) | ||
| 2-OH | 9.22, s | 9.22, s | ||
| 4-OH | 9.22, s | 9.22, s | ||
| 14-OH | 4.80, d (5.1) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Y.; Tan, Q.; Wu, J.; Chen, T.; Yang, W.; She, Z.; Wang, B. The Polyketides with Antimicrobial Activities from a Mangrove Endophytic Fungus Trichoderma lentiforme ML-P8-2. Mar. Drugs 2023, 21, 566. https://doi.org/10.3390/md21110566
Yin Y, Tan Q, Wu J, Chen T, Yang W, She Z, Wang B. The Polyketides with Antimicrobial Activities from a Mangrove Endophytic Fungus Trichoderma lentiforme ML-P8-2. Marine Drugs. 2023; 21(11):566. https://doi.org/10.3390/md21110566
Chicago/Turabian StyleYin, Yihao, Qi Tan, Jianying Wu, Tao Chen, Wencong Yang, Zhigang She, and Bo Wang. 2023. "The Polyketides with Antimicrobial Activities from a Mangrove Endophytic Fungus Trichoderma lentiforme ML-P8-2" Marine Drugs 21, no. 11: 566. https://doi.org/10.3390/md21110566
APA StyleYin, Y., Tan, Q., Wu, J., Chen, T., Yang, W., She, Z., & Wang, B. (2023). The Polyketides with Antimicrobial Activities from a Mangrove Endophytic Fungus Trichoderma lentiforme ML-P8-2. Marine Drugs, 21(11), 566. https://doi.org/10.3390/md21110566