n-Tuples on Scaffold Diversity Inspired by Drug Hybridisation to Enhance Drugability: Application to Cytarabine
Abstract
:1. Introduction
2. Results and Discussion
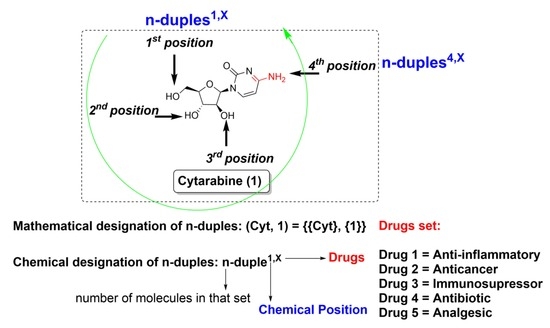
2.1. Nomenclature Foundations
2.2. Chemistry
2.2.1. Selection of Commercial Drugs
2.2.2. Syntheses of Selected Duples
2.2.3. Duples Using Linkers
3. Materials and Methods
3.1. Mass Spectrometric Analysis
3.2. Typical Protocol for Amidation
3.3. Alternative Protocol for Amidation
3.4. Protection of Cytarabine’s Primary Alcohol
3.5. Synthesis of Cytarabine-Bortezomib
3.6. Protocol for Introduction of Linker and Further Bimolecular Nucleophilic Substitution
3.7. Typical Protocol for Esterification
3.8. Principal Moments of Inertia (PMI)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Drugs Set Designation Numbers in Duples Nomenclature
| [Compound 4]: | Ibuprofen bond in position 4 | = | duple4,1 |
| [Compound 5]: | Flurbiprofen bond in position 4 | = | duple4,2 |
| [Compound 6]: | Folic acid bond in position 4 | = | duple4,3 |
| [Compound 7]: | Sulfasalazine bond in position 4 | = | duple4,4 |
| [Compound 8]: | Bortezomib bond in positions 2 and 3 | = | duple2−3,5 |
| [Compound 9]: | Methotrexate bond in position 4 | = | duple4,6 |
| [Compound 10]: | Tazobactam bond in position 4 | = | duple4,7 |
| [Compound 11]: | Ciprofloxacin bond in position 4 | = | duple4,8 |
| [Compound 12]: | Dapsone bond in position 4 | = | duple4,9 |
| [Compound 13]: | Metoclopramide bond in position 4 | = | duple4,10 |
| [Compound 14]: | Hydroxychloroquine bond in position 4 | = | duple4,11 |
| [Compound 15]: | Norfloxacin bond in position 4 | = | duple4,12 |
| [Compound 16]: | Furosemide bond in position 4 | = | duple4,13 |
| [Compound 17]: | Cilastatin bond in position 4 | = | duple4,14 |
| [Compound 18]: | Alprostadil bond in position 4 | = | duple4,15 |
| [Compound 19]: | Desamethasone bond in position 4 | = | duple4,16 |
| [Compound 23]: | Flurbiprofen bond in position 1 | = | duple1,2 |
| [Compound 4]: | Ibuprofen bond in position 4 | = | Cyt4,1 |
| [Compound 5]: | Flurbiprofen bond in position 4 | = | Cyt4,2 |
| [Compound 6]: | Folic acid bond in position 4 | = | Cyt4,3 |
| [Compound 7]: | Sulfasalazine bond in position 4 | = | Cyt4,4 |
| [Compound 8]: | Bortezomib bond in positions 2 and 3 | = | Cyt2−3,5 |
| [Compound 9]: | Methotrexate bond in position 4 | = | Cyt4,6 |
| [Compound 10]: | Tazobactam bond in position 4 | = | Cyt4,7 |
| [Compound 11]: | Ciprofloxacin bond in position 4 | = | Cyt4,8 |
| [Compound 12]: | Dapsone bond in position 4 | = | Cyt4,9 |
| [Compound 13]: | Metoclopramide bond in position 4 | = | Cyt4,10 |
| [Compound 14]: | Hydroxychloroquine bond in position 4 | = | Cyt4,11 |
| [Compound 15]: | Norfloxacin bond in position 4 | = | Cyt4,12 |
| [Compound 16]: | Furosemide bond in position 4 | = | Cyt4,13 |
| [Compound 17]: | Cilastatin bond in position 4 | = | Cyt4,14 |
| [Compound 18]: | Alprostadil bond in position 4 | = | Cyt4,15 |
| [Compound 19]: | Desamethasone bond in position 4 | = | Cyt4,16 |
| [Compound 23]: | Flurbiprofen bond in position 1 | = | Cyt1,2 |
References
- Garcia-Castro, M.; Kremer, L.; Reinkemeier, C.D.; Unkelbach, C.; Strohmann, C.; Ziegler, S.; Ostermann, C.; Kumar, K. De novo branching cascades for structural and functional diversity in small molecules. Nat. Commun. 2015, 6, 6516. [Google Scholar] [CrossRef] [PubMed]
- Vogt, M. Exploring chemical space—Generative models and their evaluation. Artif. Int. Life Sci. 2023, 3, 100064. [Google Scholar] [CrossRef]
- Villar, H.O.; Hansen, M.R. Design of chemical libraries for screening. Expert Opin. Drug Discov. 2009, 4, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S. Diversity-oriented synthesis: Exploring the intersections between chemistry and biology. Nat. Chem. Biol. 2005, 1, 74–84. [Google Scholar] [CrossRef]
- Kumar, K.; Waldmann, H. Synthesis of natural product inspired compound collections. Angew. Chem. Int. Ed. Engl. 2009, 48, 3224–3242. [Google Scholar] [CrossRef]
- Burke, M.D.; Schreiber, S.L. A planning strategy for diversity-oriented synthesis. Angew. Chem. Int. Ed. Engl. 2004, 43, 46–58. [Google Scholar] [CrossRef]
- Garcia-Castro, M.; Zimmermann, S.; Sankar, M.G.; Kumar, K. Scaffold diversity synthesis and its application in probe and drug discovery. Angew. Chem. Int. Ed. Engl. 2016, 55, 7586–7605. [Google Scholar] [CrossRef]
- Fazeela Mahaboob Begum, S.M.; Hemalatha, S. Marine Natural Products—A vital source of novel biotherapeutics. Curr. Pharm. Rep. 2022, 8, 339–349. [Google Scholar] [CrossRef]
- Lemos, T.; Merchant, A. The hedgehog pathway in hematopoiesis and hematological malignancy. Front. Oncol. 2022, 12, 960943. [Google Scholar] [CrossRef]
- Jimenez, P.C.; Wilke, D.V.; Costa-Lotufo, L.V. Marine drugs for cancer: Surfacing biotechnological innovations from the oceans. Clinics 2018, 73, e482s. [Google Scholar] [CrossRef]
- Barreca, M.; Spanò, V.; Montalbano, A.; Cueto, M.; Díaz Marrero, A.R.; Deniz, I.; Erdoğan, A.; Bilela, L.L.; Moulin, C.; Taffin-de-Givenchy, E.; et al. Marine anticancer agents: An overview with particular focus on their chemical classes. Mar. Drugs 2020, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Mieczkowski, A.; Makowska, M.; Sekula, J.; Tomczyk, E.; Zalewska, E.; Nasulweicz-Goldeman, A.; Wietrzyk, J. Bicyclic cytarabine analogues: Synthesis and investigation of antitumor properties of novel, 6-aryl-and 6-alkyl-3H-pyrrolo[2,3-d] pyrimidin-2(7H)-one arabinosides. Tetrahedron 2015, 71, 8454–8461. [Google Scholar] [CrossRef]
- Güngör, Ö.; Ҫeşme, M.; Ҫinar, M.E.; Gölcü, A. The new metal-based compound from anticancer drug cytarabine: Spectral, electrochemical, DNA-binding, antiproliferative effect and in silico studies. J. Mol. Struct. 2019, 1193, 532–543. [Google Scholar] [CrossRef]
- Sharma, P.; Dube, B.; Sawant, K. Synthesis of cytarabine lipid drug conjugate for treatment of meningeal leukemia: Development, characterization and in vitro cell line studies. J. Biom. Nanotech. 2012, 8, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Bazzanini, R.; Gouy, M.H.; Peyrottes, S.; Gosselin, G.; Périgaud, C.; Manfredini, S. Synthetic approaches to a mononucleotide prodrug of cytarabine. Nucleosides Nucleotides Nucleic Acids 2005, 24, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Najafi, M.D.; Kowsari, E.; Naderi, H.R.; Tafreshi, S.S.; Chinnappan, A.; Ramakrishna, S.; de Leew, N.H.; Ehsani, A. High-performance symmetric supercapacitor based on new functionalized graphene oxide composites with pyrimidine nucleotide and nucleoside. J. Mol. Liq. 2022, 348, 118381. [Google Scholar] [CrossRef]
- Wang, F.; Cui, C.; Ren, Z.; Wang, L.; Liu, H.; Cui, G. Preparation and biological evaluation of tumor-specific Ara-C liposomal preparations containing RGDV motif. J. Pharm. Sci. 2012, 101, 4559–4568. [Google Scholar] [CrossRef]
- D’Angelo, J.P.; West, D.B. Mathematical Thinking/Problem-Solving and Proofs, 2nd ed.; Prentice-Hall: Hoboken, NJ, USA, 2000; ISBN 978-93-5343-309-3. [Google Scholar]
- Devlin, K. The Joy of Sets. Fundamentals of Contemporary Set Theory, 2nd ed.; Ewing, J.H., Halmos, P.R., Gehring, F.W., Eds.; Springer: New York, NY, USA, 1993; pp. 7–9. ISBN 978-03-8794-094-6. [Google Scholar]
- Yu, F.; Tu, Y.; Luo, S.; Xiao, X.; Yao, W.; Jiang, M.; Jiang, X.; Yang, R.; Yuan, Y. Dual-drug backboned polyprodrug with a predefined drug combination for synergestic chemotherapy. Nano Lett. 2021, 21, 2216–2223. [Google Scholar] [CrossRef]
- Katmerlikaya, T.G.; Dag, A.; Ozgen, P.S.O.; Ersen, B.C. Dual-drug conjugated glyco-nanoassemblies for tumor-triggered targeting and synergistic cancer therapy. ACS Appl. Bio Mater. 2022, 5, 5336–5364. [Google Scholar] [CrossRef]
- Wu, Y.; Lv, S.; Li, Y.; He, H.; Ji, Y.; Zheng, M.; Liu, Y.; Yin, L. Co-delivery of dual chemo-drugs with precisely controlled, high drug loading polymeric micelles for synergistic anti-cancer therapy. Biomater. Sci. 2020, 8, 949–959. [Google Scholar] [CrossRef]
- Detappe, A.; Nguyen, H.V.-T.; Jiang, Y.; Agius, M.P.; Wang, W.; Mathieu, C.; Su, N.K.; Kristufek, S.L.; Lundberg, D.J.; Bhagchandani, S.; et al. Molecular bottlebrush prodrugs as mono- and triplex combination therapies for multiple myeloma. Nat. Nanotechnol. 2023, 18, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Q.; Wang, X.; Chen, L.; He, J.-X.; Miao, Z.-H.; Shen, J.-K. Novel liver-specific cholic acid-cytarabine conjugates with potent antitumor activities: Synthesis and biological characterization. Acta Pharmacol. Sin. 2011, 32, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Novotny, L.; Rauko, P. Cytarabine conjugates with biologically active molecules and their potential anticancer activity. Neoplasma 2009, 56, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Domenic, A.; Nicola, G.; Daniela, T.; Fulvio, C.; Nicola, A.; Orazio, N. De Novo design of targeted chemical libraries based on artificial intelligence and pair-based multiobjective optimization. J. Chem. Inf. Model. 2020, 60, 4582–4593. [Google Scholar] [CrossRef]
- Brown, N.; Ertl, P.; Lewis, R.; Luksch, T.; Reker, D.; Schneider, N. Artificial intelligence in chemistry and drug design. J. Comput. Aided Mol. Des. 2020, 34, 709–715. [Google Scholar] [CrossRef]
- Peek, R.M., Jr.; Mohla, S.; DuBois, R.N. Inflammation in the genesis and perpetuation of cancer: Summary and recommendations from a national cancer institute–sponsored meeting. Cancer Res. 2005, 65, 8583–8586. [Google Scholar] [CrossRef]
- Huang, M.; Stolina, M.; Sharma, S.; Mao, J.T.; Zhu, L.; Miller, P.W.; Wollman, J.; Herschman, H.; Dubinett, S.M. Non-small cell lung cancer cyclooxygenase-2-dependent regulation of cytokine balance in lymphocytes and macrophages: Up-regulation of interleukin 10 and down-regulation of interleukin 12 production. Cancer Res. 1998, 58, 1208–1216. [Google Scholar]
- Baxevanis, C.N.; Reclos, G.J.; Gritzapis, A.D.; Dedousis, G.V.Z.; Missitzis, I.; Papamichail, M. Elevated prostaglandin E2 production by monocytes is responsible for the depressed levels of natural killer and lymphokine-activated killer cell function in patients with breast cancer. Cancer 1993, 72, 491–501. [Google Scholar] [CrossRef]
- Dvorak, H.F. Tumours: Wounds that do not heal. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Coney, L.R.; Tomassetti, A.; Carayannopoulos, L.; Frasca, V.; Kamen, B.A.; Colnaghi, M.I.; Zurawski, V.R., Jr. Cloning of a tumor-associated antigen: MOv18 and MOv19 antibodies recognize a folate-binding protein. Cancer Res. 1991, 51, 6125–6132. [Google Scholar] [PubMed]
- Low, P.S.; Henne, W.A.; Doorneweerd, D.D. Discovery and development of folic-acid-based receptor targeting for imaging and therapy of cancer and inflammatory diseases. Acc. Chem. Res. 2008, 41, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Salazar, M.D.; Ratnam, M. The folate receptor: What does it promise in tissue-targeted therapeutics? Cancer Metastasis Rev. 2007, 26, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Sabharanjak, S.; Mayor, S. Folate receptor endocytosis and trafficking. Adv. Drug Deliv. Rev. 2004, 56, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Pestell, R.G.; Rizvanov, A.A. Antibiotics for cancer therapy. Oncotarget 2015, 6, 2587–2588. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Mukherjee, S. Cancer therapy using antibiotics. J. Cancer Ther. 2015, 6, 849–858. [Google Scholar] [CrossRef]
- Kochanowskia, K.; Morinishia, L.; Altschuler, S.J.; Wu, L.F. Drug persistence—From antibiotics to cancer therapies. Curr. Opin. Syst. Biol. 2018, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shang, Q.; Li, W.; Guo, W.; Stojadinovic, A.; Mannion, C.; Man, Y.-G.; Chen, T. Antibiotics for cancer treatment: A double-edged sword. J. Cancer 2020, 11, 5135–5149. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Immunosuppressants in cancer prevention and therapy. OncoImmunology 2013, 2, e26961. [Google Scholar] [CrossRef]
- Naylon, M.R.; Bockus, A.T.; Blanco, M.-J.; Lokey, R.S. Cyclic peptide natural products chart the frontier of oral bioavailability in the pursuit of undruggable targets. Curr. Opin. Chem. Biol. 2017, 38, 141–147. [Google Scholar]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 6, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Sauer, W.H.B.; Schwarz, M.K. Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J. Chem. Inf. Comput. Sci. 2003, 43, 987–1003. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Ito, T.; Fujii, H.G.; Sato, S. Synthesis and reduction kinetics of five ibuprofen-nitroxides for ascorbic acid and methyl radicals. Chem. Pharm. Bull. 2016, 64, 1509–1513. [Google Scholar] [CrossRef] [PubMed]
- Young, I.S.; Baran, P.S. Protecting-group-free synthesis as an opportunity for invention. Nat. Chem. 2009, 1, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.K. Genetic factors influencing cytarabine therapy. Pharmacogenomics 2009, 10, 1657–1674. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Yang, H.; Zheng, D.; Ye, J.; Zhou, Q.; Li, G.; Zhou, Y.; Yang, X.; Liu, J.; Ye, F.; et al. Smart and generalizable cytarabine derivative-triggered nanoparticles for synergistic therapy of relapsed/refractory acute myeloid leukemia. ACS Appl. Mater. Interfaces 2023, 15, 27624–27637. [Google Scholar] [CrossRef] [PubMed]
- Liao, A.-M.; Cai, B.; Huang, J.-H.; Hui, M.; Lee, K.-K.; Lee, K.Y.; Chun, C. Synthesis, anticancer activity and potential application of diosgenin modified cancer chemotherapeutic agent cytarabine. Food Chem. Toxicol. 2021, 148, 111920. [Google Scholar] [CrossRef]
- Sarabia, F.; García-Castro, M.; Chammaa, S.; Sánchez-Ruíz, A. The chiron approach to pironetins: Synthesis of the δ-lactonic fragment and modified building blocks from D-glucal. J. Carb. Chem. 2006, 25, 267–280. [Google Scholar] [CrossRef]
- McMurry, J.E.; Dushin, R.G. Total synthesis of (±)-crassin by titanium-induced pinacol coupling. J. Am. Chem. Soc. 1989, 111, 8928–8929. [Google Scholar] [CrossRef]
- Puppala, R.; Pathi, S.L.; Rao, D.R.; Kankan, R.N. Process for the Preparation of Bortezomib Mannitol Ester. PCT/GB2O14/OOO150. U.S. Patent 2016/0075736A1, 17 March 2016. [Google Scholar]
- Wu, K.; Cheng, R.; Zhang, J.; Meng, F.; Deng, C.; Zhong, Z. Micellar nanoformulation of lipophilized Bortezomib: High drug loading, improved tolerability and targeted treatment of triple negative breast cancer. J. Mater. Chem. B 2017, 5, 5658–5667. [Google Scholar] [CrossRef]
- Berrío Escobar, J.F.; Pastrana Restrepo, M.H.; Galeano Jaramillo, E.; Márquez Fernández, D.M.; Márquez Fernández, M.E.; Martínez Martínez, A. Synthesis and cytotoxic activity of per-acetylated and halogenated derivatives of nucleosides in breast cancer cells. Ars Pharm. 2017, 58, 145–154. [Google Scholar] [CrossRef]
- Marsch, A.; Khan, A.; Haddleton, D.M.; Hannon, M.J. Atom transfer polymerization: Use of uridine and adenosine derivatized monomers and initiatiors. Macromolecules 1999, 32, 8725–8731. [Google Scholar] [CrossRef]
- Ferri, M.; Alunno, M.; Greco, F.A.; Mammoli, A.; Saluti, G.; Carotti, A.; Sardella, R.; Macchiarulo, A.; Camaioni, E.; Liscio, P. Fragment based drug design and diversity-oriented synthesis of carboxylic acid isosteres. Bioorg. Med. Chem. 2020, 28, 115731. [Google Scholar] [CrossRef] [PubMed]
- Gilham, D.; Lehner, R. Techniques to measure lipase and esterase activity in vitro. Methods 2005, 36, 139–147. [Google Scholar] [CrossRef]
- Iavorschi, M.; Lupăescu, A.-V.; Darie-Ion, L.; Indeykina, M.; Hitruc, G.E.; Petre, B.A. Cu and Zn Interactions with Peptides Revealed by High-Resolution Mass Spectrometry. Pharmaceuticals 2022, 15, 1096. [Google Scholar] [CrossRef]
- Zieba, A.; Maslankiewicz, A.; Sitkowski, J. 1H, 13C and 15N NMR spectra of ciprofloxacin. Magn. Reson. Chem. 2004, 42, 903–904. [Google Scholar] [CrossRef]









| General Chemical Nomenclature | Specific Chemical Nomenclature | Mathematical Nomenclature |
|---|---|---|
| n-duples1,X | n-Cyt1,X | (Cyt, 1) = {Cyt}, {Cyt, 1}; where Cyt = Central drug, in our case cytarabine, which is covalently bonded in position 1 to drug X (drug number). |
| n-duples2,X | n-Cyt2,X | (Cyt, 2) = {Cyt}, {Cyt, 2}; where Cyt = Central drug, in our case cytarabine, which is covalently bonded in position 2 to drug X (drug number). |
| n-duplesY,1 | n-CytY,1 | It is included in diverse mathematical nomenclature. Drug 1 is attached to different positions in the central drug, in our case, cytarabine. |
| Entry | Drug | Medical Uses | Main Functional Group 1 |
|---|---|---|---|
| 1 | Ibuprofen | Anti-inflammatory | Carboxylic acid |
| 2 | Flurbiprofen | Anti-inflammatory | Carboxylic acid |
| 3 | Norfloxacin | Antibiotic | Carboxylic acid |
| 4 | Hydroxychloroquine | Immunosuppressor | Carboxylic acid |
| 5 | Metoclopramide | Antiemetic | Amine |
| 6 | Furosemide | Diuretic | Carboxylic acid |
| 7 | Cilastatin | Antibiotic | Carboxylic acid |
| 8 | Alprostadil | Erectile dysfunction | Carboxylic acid |
| 9 | Ciprofloxacin | Antibiotic | Carboxylic acid |
| 10 | Sulfasalazine | Immunosuppressor | Carboxylic acid |
| 11 | Methotrexate | Anticancer | Carboxylic acid |
| 12 | Tazobactam | Antibiotic | Carboxylic acid |
| 13 | Dexamethasone | Steroid | Alcohol |
| 14 | Folic acid | Vitamin | Carboxylic acid |
| 15 | Dapsone | Antibiotic | Amine |
| 16 | Bortezomib | Anticancer | Boronic acid |
| Entry | Drug | Substitution Positions 1 | Lipinski Rules? 2,3 |
|---|---|---|---|
| 1 | Ibuprofen | 1; 2; 3; 4 | Ro5 |
| 2 | Flurbiprofen | 1; 2; 3; 4 | Ro5 |
| 3 | Norfloxacin | 1; 2; 3; 4 | bRo5 |
| 4 | Hydroxychloroquine | 1; 2; 3; 4 | Ro5 |
| 5 | Metoclopramide | 1; 2; 3; 4 | bRo5 |
| 6 | Furosemide | 1; 2; 3; 4 | bRo5 |
| 7 | Cilastatin | 1; 2; 3; 4 | bRo5 |
| 8 | Alprostadil | 1; 2; 3; 4 | bRo5 |
| 9 | Ciprofloxacin | 1; 2; 3; 4 | bRo5 |
| 10 | Sulfasalazine | 1; 2; 3; 4 | bRo5 |
| 11 | Methotrexate | 1; 2; 3; 4 | bRo5 |
| 12 | Tazobactam | 1; 2; 3; 4 | bRo5 |
| 13 | Dexamethasone | 1; 2; 3; 4 | bRo5 |
| 14 | Folic acid | 1; 2; 3; 4 | bRo5 |
| 15 | Dapsone | 1; 2; 3; 4 | Ro5 |
| 16 | Bortezomib | 1; 2; 3; 4 | bRo5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Castro, M.; Fuentes-Rios, D.; López-Romero, J.M.; Romero, A.; Moya-Utrera, F.; Díaz-Morilla, A.; Sarabia, F. n-Tuples on Scaffold Diversity Inspired by Drug Hybridisation to Enhance Drugability: Application to Cytarabine. Mar. Drugs 2023, 21, 637. https://doi.org/10.3390/md21120637
García-Castro M, Fuentes-Rios D, López-Romero JM, Romero A, Moya-Utrera F, Díaz-Morilla A, Sarabia F. n-Tuples on Scaffold Diversity Inspired by Drug Hybridisation to Enhance Drugability: Application to Cytarabine. Marine Drugs. 2023; 21(12):637. https://doi.org/10.3390/md21120637
Chicago/Turabian StyleGarcía-Castro, Miguel, David Fuentes-Rios, J. Manuel López-Romero, Antonio Romero, Federico Moya-Utrera, Amelia Díaz-Morilla, and Francisco Sarabia. 2023. "n-Tuples on Scaffold Diversity Inspired by Drug Hybridisation to Enhance Drugability: Application to Cytarabine" Marine Drugs 21, no. 12: 637. https://doi.org/10.3390/md21120637
APA StyleGarcía-Castro, M., Fuentes-Rios, D., López-Romero, J. M., Romero, A., Moya-Utrera, F., Díaz-Morilla, A., & Sarabia, F. (2023). n-Tuples on Scaffold Diversity Inspired by Drug Hybridisation to Enhance Drugability: Application to Cytarabine. Marine Drugs, 21(12), 637. https://doi.org/10.3390/md21120637