A Survey of Didemnin Depsipeptide Production in Tistrella
Abstract
:1. Introduction

2. Results
2.1. Establishing Viable Culture Conditions
2.2. Comparative Analyses for the Production of Didemnin Depsipeptides
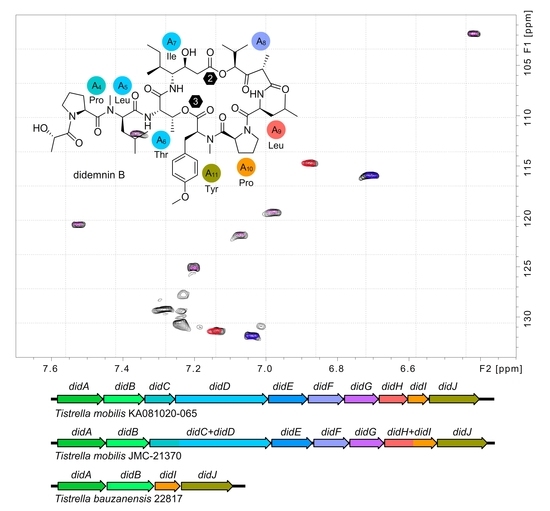
2.3. NMR Data Collection
2.4. Atomic Sort Metabolomics Analysis
2.5. Didemnin BGC Architecture Analysis
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Culture Conditions
4.3. Extraction
4.4. NMR Analyses
4.5. Purification of Didemnin B
4.6. Genome Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rinehart, K.L., Jr.; Gloer, J.B.; Hughes, R.G., Jr.; Renis, H.E.; McGovren, J.P.; Swynenberg, E.B.; Stringfellow, D.A.; Kuentzel, S.L.; Li, L.H. Didemnins: Antiviral and antitumor depsipeptides from a caribbean tunicate. Science 1981, 212, 933–935. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, H.; Fenical, W.; Epifanio, R.D.A. Tamandarins A and B: New Cytotoxic Depsipeptides from a Brazilian Ascidian of the Family Didemnidae. J. Org. Chem. 2000, 65, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Currano, J.N.; Carroll, P.J.; Joullié, M.M. Didemnins, tamandarins and related natural products. Nat. Prod. Rep. 2012, 29, 404–424. [Google Scholar] [CrossRef] [PubMed]
- Vera, M.D.; Joullié, M.M. Natural products as probes of cell biology: 20 years of didemnin research. Med. Res. Rev. 2002, 22, 102–145. [Google Scholar] [CrossRef] [PubMed]
- Le Tourneau, C.; Raymond, E.; Faivre, S. Aplidine: A Paradigm of how to Handle the Activity and Toxicity of a Novel Marine Anticancer Poison. Curr. Pharm. Des. 2007, 13, 3427–3439. [Google Scholar] [CrossRef]
- Leisch, M.; Egle, A.; Greil, R. Plitidepsin: A potential new treatment for relapsed/refractory multiple myeloma. Future Oncol. 2019, 15, 109–120. [Google Scholar] [CrossRef]
- Alonso-Álvarez, S.; Pardal, E.; Sánchez-Nieto, D.; Navarro, M.; Caballero, M.D.; Mateos, M.V.; Martin, A. Plitidepsin: Design, development, and potential place in therapy. Drug Des. Dev. Ther. 2017, 11, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Chun, H.G.; Davies, B.; Hoth, D.; Suffness, M.; Plowman, J.; Flora, K.; Grieshaber, C.; Leyland–Jones, B. Didemnin B. The first marine compound entering clinical trials as an antineoplastic agent. Investig. New Drugs. 1986, 4, 279–284. [Google Scholar] [CrossRef]
- Weed, S.D.; Stringfellow, D.A. Didemnins A and B. Effectiveness against cutaneous herpes simplex virus in mice. Antivir. Res. 1983, 3, 269–274. [Google Scholar] [CrossRef]
- Rodon, J.; Muñoz–Basagoiti, J.; Perez–Zsolt, D.; Noguera–Julian, M.; Paredes, R.; Mateu, L.; Quiñones, C.; Perez, C.; Erkizia, I.; Blanco, I.; et al. Identification of Plitidepsin as Potent Inhibitor of SARS–CoV–2–Induced Cytopathic Effect After a Drug Repurposing Screen. Front. Pharmacol. 2021, 12, 646676. [Google Scholar] [CrossRef]
- Sachse, M.; Tenorio, R.; Fernández de Castro, I.; Muñoz–Basagoiti, J.; Perez–Zsolt, D.; Raïch–Regué, D.; Rodon, J.; Losada, A.; Avilés, P.; Cuevas, C.; et al. Unraveling the antiviral activity of plitidepsin against SARS–CoV–2 by subcellular and morphological analysis. Antiviral. Res. 2022, 200, 105270. [Google Scholar] [CrossRef] [PubMed]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L.; et al. Plitidepsin has potent preclinical efficacy against SARS–CoV–2 by targeting the host protein eEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Varona, J.F.; Landete, P.; Lopez–Martin, J.A.; Estrada, V.; Paredes, R.; Guisado–Vasco, P.; Fernandez de Orueta, L.; Torralba, M.; Fortun, J.; Vates, R.; et al. Preclinical and randomized phase I studies of plitidepsin in adults hospitalized with COVID–19. Life Sci. Alliance 2022, 5, e202101200. [Google Scholar] [CrossRef] [PubMed]
- Tsukimoto, M.; Nagaoka, M.; Shishido, Y.; Fujimoto, J.; Nishisaka, F.; Matsumoto, S.; Harunari, E.; Imada, C.; Matsuzaki, T. Bacterial Production of the Tunicate-Derived Antitumor Cyclic Depsipeptide Didemnin B. J. Nat. Prod. 2011, 74, 2329–2331. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.H.; Arunpairojana, V.; Palakawong, S.; Yokota, A. Tistrella mobilis gen. nov., sp. nov., a novel polyhydroxyalkanoate–producing bacterium belonging to alpha–Proteobacteria. J. Gen. Appl. Microbiol. 2002, 48, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.-C.; Liu, H.-C.; Zhou, Y.-G.; Schinner, F.; Margesin, R. Tistrella bauzanensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 2227–2230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Kersten, R.D.; Nam, S.-J.; Lu, L.; Al-Suwailem, A.M.; Zheng, H.; Fenical, W.; Dorrestein, P.C.; Moore, B.S.; Qian, P.-Y. Bacterial Biosynthesis and Maturation of the Didemnin Anti-cancer Agents. J. Am. Chem. Soc. 2012, 134, 8625–8632. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; van der Donk, W.A. Biosynthesis of 3–thia–α–amino acids on a carrier peptide. Proc. Natl. Acad. Sci. USA 2022, 119, e2205285119. [Google Scholar] [CrossRef]
- Lu, L.; Meehan, M.J.; Gu, S.; Chen, Z.; Zhang, W.; Zhang, G.; Liu, L.; Huang, X.; Dorrestein, P.C.; Xu, Y.; et al. Mechanism of Action of Thalassospiramides, A New Class of Calpain Inhibitors. Sci. Rep. 2015, 5, 8783. [Google Scholar] [CrossRef] [Green Version]
- Ross, A.C.; Xu, Y.; Lu, L.; Kersten, R.D.; Shao, Z.; Al-Suwailem, A.M.; Dorrestein, P.C.; Qian, P.-Y.; Moore, B.S. Biosynthetic Multitasking Facilitates Thalassospiramide Structural Diversity in Marine Bacteria. J. Am. Chem. Soc. 2013, 135, 1155–1162. [Google Scholar] [CrossRef]
- Duggan, B.M.; Cullum, R.; Fenical, W.; Amador, L.A.; Rodríguez, A.D.; La Clair, J.J. Searching for Small Molecules with an Atomic Sort. Angew. Chem. Int. Ed. 2020, 59, 1144–1148. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Sünninghausen, D.; Becker, J.; Luy, B. Rapid Heteronuclear Single Quantum Correlation NMR Spectra at Natural Abundance. J. Am. Chem. Soc. 2014, 136, 1242–1245. [Google Scholar] [CrossRef] [PubMed]
- bcl2fastq: A Proprietary Illumina Software for the Conversion of bcl Files to Basecalls. Available online: https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html (accessed on 11 January 2023).
- An Open Source Software for the QC and Adapter Trimming of ONT Technologies. Available online: https://github.com/rrwick/Porechop (accessed on 11 January 2023).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]



| Antibiotic (Concentration) | T. mobilis JCM21370 | T. mobilis YIT12409 |
|---|---|---|
| apramycin (50 µg/mL) | − | − |
| carbenicillin (50 µg/mL) | +++ | +++ |
| chloramphenicol (25 µg/mL) | − | − |
| colistin (5 µg/mL) | +++ | − |
| kanamycin (50 µg/mL) | − | − |
| hygromycin (100 µg/mL) | − | − |
| nalidixic acid (30 µg/mL) | + | − |
| none | +++ | +++ |
| Culture | Strain a | Media b | V (mL) | T (°C) | Days |
|---|---|---|---|---|---|
| A | JCM21370 | 74NB | 200 | 30 | 2 |
| B | JCM21370 | 74NB | 5000 | 30 | 3 |
| C | YIT12409 | 74NB | 1000 | 30 | 3 |
| D | YIT12409 | TSB | 1000 | 30 | 3 |
| E | JCM21370 | 74NB | 100 | 30 | 3 |
| F | YIT12409 | 74NB | 100 | 30 | 3 |
| G | DSM22817 | 74NB | 100 | 30 | 10 |
| H | JCM21370 | 74NB | 2000 | 30 | 3 |
| I | YIT12409 | 74NB | 2000 | 30 | 3 |
| J | DSM22817 | 74NB | 2000 | 20 | 6 |
| K | JCM21370 | 74NB | 1000 | 30 | 3 |
| L | YIT12409 | 74NB | 1000 | 30 | 3 |
| M | JCM21370 | 74NB | 1000 | 30 | 3 |
| N | JCM21370 | GYP | 1000 | 30 | 3 |
| O | JCM21370 | SBM | 1000 | 30 | 3 |
| P | YIT12409 | 74NB | 1000 | 30 | 3 |
| Q | YIT12409 | GYP | 1000 | 30 | 3 |
| R | YIT12409 | SBM | 1000 | 30 | 3 |
| S | JCM21370 | GYP | 2000 | 30 | 3 |
| Compound | T. mobilis JCM21370 (H a) | T. bauzanensis DSM22817 (I a) | T. mobilis YIT12409 (J a) |
|---|---|---|---|
| methanol | 0.009 b, 1/1 c | 0.001, 1/1 | 0.002, 1/1 |
| indole | 0.032, 6/6 | 0.095, 6/6 | 0.058, 6/6 |
| didemnin B (1) | 0.056, 51/51 | 0.587, 5/51 d | 0.389, 23/51 |
| cyclo(Pro–Tyr) (3) | 0.063, 11/11 | 0.044, 11/11 | 0.142, 10/11 |
| Leu–Pro–Ile–Pro–Ile (2) | 0.074, 27/27 | 0.054, 26/27 | 0.101, 23/27 |
| cyclo(Ile–Pro–Leu–Pro) | 0.183, 19/26 | 0.175, 21/26 | 0.154, 17/26 |
| n–hexane | 0.136, 2/3 | 0.123, 3/3 | 0.121, 2/3 |
| Database Hit Compound | T. mobilis JCM21370 (H a) | T. bauzanensis DSM22817 (I a) | T. mobilis YIT12409 (J a) | |||
|---|---|---|---|---|---|---|
| [] b, mM | Yield c, µg | [] b, mM | Yield c, µg | [] b, mM | Yield c, µg | |
| indole | 51.2 | 300.7 | 9.0 | 53.1 | 135.4 | 795.5 |
| didemnin B (1) | 7.7 | 427.5 | – | – | 1.8 | 102.7 |
| cyclo(Pro–Tyr) (3) | 22.6 | 294.2 | 25.6 | 333.2 | 22.7 | 295.9 |
| Leu–Pro–Ile–Pro–Ile (2) | 17.5 | 483.5 | 18.4 | 507.5 | 16.3 | 449.9 |
| cyclo(Ile–Pro–Leu–Pro) | 30.6 | 643.9 | 25.7 | 540.2 | 28.0 | 588.9 |
| Media | T. mobilis JCM21370 | T. mobilis YIT12409 | ||||||
|---|---|---|---|---|---|---|---|---|
| Culture a | [] b, mM | Yield c, µg | Titer d, mg/L | Culture a | [] b, mM | Yield c, µg | Titer d, mg/L | |
| 74NB | K | 7.1 | 397.3 | 54.8 | L | 4.4 | 247.4 | 34.1 |
| 74NB | M | 14.6 | 809.9 | 111.7 | P | 4.7 | 258.8 | 35.7 |
| GYP | N | 3.7 | 203.5 | 28.2 | Q | 6.8 | 375.8 | 51.8 |
| SBM | O | 7.1 | 393.0 | 54.2 | R | 5.7 | 319.0 | 44.0 |
| Position | δ 1H, Mult (J in Hz) | δ13C | 1H, 1H–COSY | 1H, 13C–HMBC |
|---|---|---|---|---|
| Ist1 | – | 172.9 | – | – |
| Ist2a | 3.61, m | 40.3 | Ist2b | Ist1 |
| Ist2b | 2.43, dd (10.4, 17.5) | Ist2a, Ist3 | Ist1, Ist3 | |
| Ist3 | 4.05, t (10.5) | 67.6 | Ist2b | Ist1, Ist4 |
| Ist4 | 3.97, td (10.3, 2.5) | 56.6 | IstNH, Ist5 | Ist3, Ist6 |
| Ist5 | 2.00, m | 34.9 | Ist4, Ist6 | Ist7w |
| Ist6 | 0.98, d (7.2) | 14.5 | Ist5, Ist7a, Ist7b | Ist4, Ist7 |
| Ist7a | 1.40, m | Ist5, Ist6, Ist7b, Ist8 | Ist4w, Ist5w Ist6w, Ist8w | |
| Ist7b | 1.26, m | Ist5, Ist6, Ist7a, Ist8 | Ist4w, Ist5w, Ist6w, Ist8w | |
| Ist8 | 0.97, d (6.6) | 12.1 | Ist7 | – |
| IstNH | 7.35, d (10.0) | – | Ist4 | Thr1w |
| Hip1 | – | 171.3 | – | – |
| Hip2 | 4.12, q (6.7) | 49.2 | Hip3 | Hip1, Hip3, Hip4 |
| Hip3 | 1.30, d (6.9) | 15.9 | Hip2 | Hip1, Hip2, Hip4 |
| Hip4 | – | 205.7 | – | – |
| Hip5 | 5.05, d (4.0) | 81.9 | Hip6 | Hip4, Hip6, Hip7, Hip8w, Ist1 |
| Hip6 | 2.32, m | 31.0 | Hip5, Hip7, Hip8 | Hip8w |
| Hip7 | 0.87, d (6.9) | 17.0 | Hip6 | Hip5, Hip6, Hip8 |
| Hip8 | 0.93, d (6.8) | 19.2 | Hip6 | Hip5, Hip6, Hip7 |
| Leu1 | – | 171.5 | – | – |
| Leu2 | 4.78, m | 50.6 | LeuNH, Leu3a, Leu3b | – |
| Leu3a | 1.64, m | 42.2 | Leu2, Leu3b | – |
| Leu3b | 1.23, m | Leu2, Leu3a | – | |
| Leu4 | 1.52, m | 25.7 | Leu3a, Leu3b, Leu5, Leu6 | – |
| Leu5 | 0.92, d (6.7) | 21.2 | Leu4 | Leu3, Leu4, Leu6 |
| Leu6 | 0.96, d (6.9) | 23.8 | Leu4 | Leu3, Leu4, Leu5 |
| LeuNH | 8.1, d (9.2) | – | Leu2 | Hip1 |
| Pro1 | – | 172.5 | – | – |
| Pro2 | 4.80, m | 58.4 | Pro3a, Pro3b | – |
| Pro3a | 2.19, m | 28.7 | Pro2, Pro3b | Pro1, Pro2w |
| Pro3b | 1.70, dt (12.3, 6.1) | Pro2, Pro3a | Pro1, Pro5 | |
| Pro4 | 2.06, m | 25.5 | Pro5a, Pro5b | Pro2, Pro5 |
| Pro5a | 3.74, dt (9.7, 6.8) | 47.9 | Pro4 | – |
| Pro5b | 3.58, dt (5.3, 3.1) | Pro4 | Pro1 | |
| MTyr1 | – | 170.0 | – | – |
| MTyr2 | 4.01. dd (10.6, 4.7) | 66.3 | MTyr3a, MTyr3b | MTyr1, MTyr3, MTyrNM, Pro1 |
| MTyr3a | 3.15, dd (14.1, 10.7) | 34.6 | MTyr2, MTyr3b | MTyr1w, MTyr2, MTyr4, MTyr5 |
| MTyr3b | 3.29, m | MTyr2, MTyr3a | MTyr2, MTyr4, MTyr5 | |
| MTyr4 | – | 130.6 | – | – |
| MTyr5 | 7.14, d (8.6) | 131.8 | MTyr6 | MTyr3, MTyr5, MTyr6, MTyr7 |
| MTyr6 | 6.88, d (8.5) | 114.7 | MTyr5 | MTry4, MTyr6, MTyr7 |
| MTyr7 | – | 159.8 | – | – |
| MTyr8 | 3.78, s | 55.3 | MTyr7 | |
| MTyrNMe | 2.62, s | 39.1 | – | – |
| Thr1 | – | 171.1 | – | – |
| Thr2 | 4.54, dd (5.1, 2.4) | 59.1 | ThrNH | Thr3, Thr4w |
| Thr3 | 5.37, d (5.9) | 71.1 | Thr4 | Thr2w, MTyr1 |
| Thr4 | 1.35, d (6.3) | 16.6 | Thr3 | Thr2, Thr3 |
| ThrNH | 7.94, d (4.9) | – | Thr2 | MLeu1 |
| MLeu1 | – | 173.0 | – | – |
| MLeu2 | 5.39, d (8.2) | 55.9 | MLeu3 | HPro1, MLeu1, MLeu3, MeLeu4, MLeuNMe |
| MLeu3 | 1.80, dd (8.5, 6.9) | 37.0 | MLeu2, MLeu4 | HPro1, MLeu1, MLeu2, MLeu4, MLeu5, MLeu6 |
| MLeu4 | 1.47, m | 25.5 | MLeu3, MLeu5, MLeu6 | MLeu3, MLeu4, MLeu5 |
| MLeu5 | 0.89, d (6.5) | 21.3 | MLeu4 | MLeu3, MLeu4, MLeu6 |
| MLeu6 | 0.95, d (6.7) | 23.8 | MLeu4 | MLeu3, MLeu4, MLeu5 |
| MLeuNMe | 3.19, s | 31.5 | – | MLeu2, HPro1 |
| HPro1 | – | 175.4 | – | – |
| HPro2 | 4.84, m | 58.0 | HPro3a, HPro3b | HPro3 |
| HPro3a | 2.28, m | 29.2 | HPro2, HPro3b | HPro5w |
| HPro3b | 1.85, m | HPro2, HPro3a | HPro1w | |
| HPro4a | 2.16, m | 26.6 | HPro4b, HPro5a, HPro5b | – |
| HPro4b | 2.03, m | HPro4a, HPro5a, HPro5b | HPro2, HPro3, HPro5 | |
| HPro5a | 3.88, ddd (10.8, 7.3, 3.6) | 48.1 | HPro4a, HPro4b, HPro5b | HPro4w |
| HPro5b | 3.64, dd (10.0, 6.6) | HPro4a, HPro4b, HPro5a | – | |
| HP1 | – | 174.1 | – | – |
| HP2 | 4.47, q (6.6) | 67.3 | HP3 | HP1, HP3 |
| HP3 | 1.38, d (6.6) | 19.9 | HP2 | HP1, HP2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stankey, R.J.; Johnson, D.; Duggan, B.M.; Mead, D.A.; La Clair, J.J. A Survey of Didemnin Depsipeptide Production in Tistrella. Mar. Drugs 2023, 21, 56. https://doi.org/10.3390/md21020056
Stankey RJ, Johnson D, Duggan BM, Mead DA, La Clair JJ. A Survey of Didemnin Depsipeptide Production in Tistrella. Marine Drugs. 2023; 21(2):56. https://doi.org/10.3390/md21020056
Chicago/Turabian StyleStankey, Robert J., Don Johnson, Brendan M. Duggan, David A. Mead, and James J. La Clair. 2023. "A Survey of Didemnin Depsipeptide Production in Tistrella" Marine Drugs 21, no. 2: 56. https://doi.org/10.3390/md21020056
APA StyleStankey, R. J., Johnson, D., Duggan, B. M., Mead, D. A., & La Clair, J. J. (2023). A Survey of Didemnin Depsipeptide Production in Tistrella. Marine Drugs, 21(2), 56. https://doi.org/10.3390/md21020056