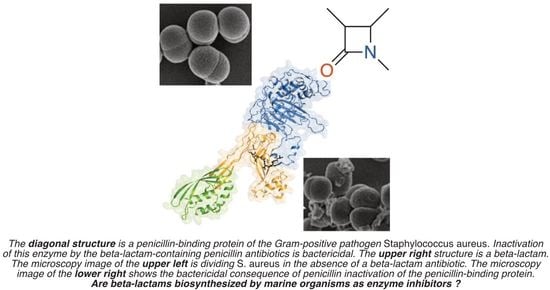
β-Lactams from the Ocean
Abstract
:1. Introduction
2. Do Marine Organisms Biosynthesize Exceptional Inhibitors of Bacterial Cell-Wall Biosynthesis?
3. Do Marine Organisms Biosynthesize The Classical Antibacterial β-Lactams?
4. Do Marine Organisms Biosynthesize β-lactams?
5. Does the Marine Environment Contain β-lactam-degrading Enzymes?
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kardos, N.; Demain, A.L. Penicillin: The medicine with the greatest impact on therapeutic outcomes. Appl. Microbiol. Biotechnol. 2011, 92, 677–687. [Google Scholar] [CrossRef]
- Ramírez-Rendon, D.; Passari, A.K.; Ruiz-Villafán, B.; Rodríguez-Sanoja, R.; Sánchez, S.; Demain, A.L. Impact of novel microbial secondary metabolites on the pharma industry. Appl. Microbiol. Biotechnol. 2022, 106, 1855–1878. [Google Scholar] [CrossRef]
- Walsh, C.T.; Wencewicz, T.A. Prospects for new antibiotics: A molecule-centered perspective. J. Antibiot. 2014, 67, 7–22. [Google Scholar] [CrossRef]
- Page, J.E.; Walker, S. Natural products that target the cell envelope. Curr. Opin. Microbiol. 2021, 61, 16–24. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez, C. Marine natural products in medicinal chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, X.; Xie, X.; Chen, B.; Liu, L.; Jiang, C.; Qian, Q. Marine natural products: A potential source of anti-hepatocellular carcinoma drugs. J. Med. Chem. 2021, 64, 7879–7899. [Google Scholar] [CrossRef]
- Wender, P.A.; Quiroz, R.V.; Stevens, M.C. Function through synthesis-informed design. Acc. Chem. Res. 2015, 48, 752–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figuerola, B.; Avila, C. The phylum Bryozoa as a promising source of anticancer drugs. Mar. Drugs 2019, 17, 477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.; Chen, H.; Chang, N.; Xu, Y.; Jiao, J.; Zhang, H. Unlocking the drug potential of the bryostatin family: Recent advances in product synthesis and biomedical applications. Chem. Eur. J. 2020, 26, 1166–1195. [Google Scholar] [CrossRef]
- Wender, P.A.; Sloane, J.L.; Luu-Nguyen, Q.H.; Ogawa, Y.; Shimizu, A.J.; Ryckbosch, S.M.; Tyler, J.H.; Hardman, C. Function-oriented synthesis: Design, synthesis, and evaluation of highly simplified bryostatin analogues. J. Org. Chem. 2020, 85, 15116–15128. [Google Scholar] [CrossRef]
- Abramson, E.; Hardman, C.; Shimizu, A.J.; Hwang, S.; Hester, L.D.; Snyder, S.H.; Wender, P.A.; Kim, P.M.; Kornberg, M.D. Designed PKC-targeting bryostatin analogs modulate innate immunity and neuroinflammation. Cell Chem. Biol. 2021, 28, 537–545. [Google Scholar] [CrossRef]
- Jackson, S.A.; Crossman, L.; Almeida, E.L.; Margassery, L.M.; Kennedy, J.; Dobson, A.D.W. Diverse and abundant secondary metabolism biosynthetic gene clusters in the genomes of marine sponge derived Streptomyces spp. isolates. Mar. Drugs 2018, 16, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tischler, D. A perspective on enzyme inhibitors from marine organisms. Mar. Drugs 2020, 18, 431. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Liao, L.; Chen, B. Complete genome analysis reveals secondary metabolite biosynthetic capabilities of Streptomyces sp. R527F isolated from the Arctic Ocean. Mar. Genomics 2022, 63, 100949. [Google Scholar] [CrossRef]
- Shi, S.; Cui, L.; Zhang, K.; Zeng, Q.; Li, Q.; Ma, L.; Long, L.; Tian, X. Streptomyces marincola sp. nov., a novel marine actinomycete, and its biosynthetic potential of bioactive natural products. Front. Microbiol. 2022, 13, 860308. [Google Scholar] [CrossRef] [PubMed]
- Mlot, C. Microbiology. Antibiotics in nature: Beyond biological warfare. Science 2009, 324, 1637–1639. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Uppal, S.; Metz, J.L.; Xavier, R.K.M.; Nepal, K.K.; Xu, D.; Wang, G.; Kwan, J.C. Uncovering lasonolide A biosynthesis using genome-resolved metagenomics. mBio 2022, 13, e0152422. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Lin, Z. Translating marine symbioses toward drug development. mBio 2022, 13, e0249922. [Google Scholar] [CrossRef]
- Testero, S.A.; Llarrull, L.; Fisher, J.F.; Mobashery, S. β-Lactam antibiotics. Burg. Med. Chem. Drug Discov. Dev. 2021, 7, 1–188. [Google Scholar] [CrossRef]
- De Benedetti, S.; Fisher, J.F.; Mobashery, S. Bacterial cell wall: Morphology and biochemistry: Chapter 18. In Practical Handbook of Microbiology, 4th ed.; Taylor and Francis: New York, NY, USA, 2021; pp. 167–204. [Google Scholar] [CrossRef]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [Green Version]
- Pratt, R.F. β-Lactamases: Why and how. J. Med. Chem. 2016, 59, 8207–8220. [Google Scholar] [CrossRef]
- McCauley, E.P.; Piña, I.C.; Thompson, A.D.; Bashir, K.; Weinberg, M.; Kurz, S.L.; Crews, P. Highlights of marine natural products having parallel scaffolds found from marine-derived bacteria, sponges, and tunicates. J. Antibiot. 2020, 73, 504–525. [Google Scholar] [CrossRef]
- Bauman, K.D.; Shende, V.V.; Chen, P.Y.; Trivella, D.B.B.; Gulder, T.A.M.; Vellalath, S.; Romo, D.; Moore, B.S. Enzymatic assembly of the salinosporamide γ-lactam-β-lactone anticancer warhead. Nat Chem. Biol. 2022, 18, 538–546. [Google Scholar] [CrossRef]
- Gulder, T.A.; Moore, B.S. Salinosporamide natural products: Potent 20S proteasome inhibitors as promising cancer chemotherapeutics. Angew. Chem. Int. Ed. 2010, 49, 9346–9367. [Google Scholar] [CrossRef] [PubMed]
- Della Sala, G.; Agriesti, F.; Mazzoccoli, C.; Tataranni, T.; Costantino, V.; Piccoli, C. Clogging the ubiquitin-proteasome machinery with marine natural products: Last decade update. Mar. Drugs 2018, 16, 467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano-Aparicio, N.; Moliner, V.; Świderek, K. On the origin of the different reversible characters of salinosporamide A and homosalinosporamide A in the covalent inhibition of the human 20S proteasome. ACS Catal. 2021, 11, 11806–11819. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Gottwalt, S.; Beyer, P.; Butler, M.; Czaplewski, L.; Lienhardt, C.; Moja, L.; Paul, M.; Paulin, S.; Rex, J.H.; et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect. Dis. 2019, 19, e40–e50. [Google Scholar] [CrossRef] [PubMed]
- Chahine, E.B.; Dougherty, J.A.; Thornby, K.A.; Guirguis, E.H. Antibiotic approvals in the last decade: Are we keeping up with resistance? Ann. Pharmacother. 2022, 56, 441–462. [Google Scholar] [CrossRef]
- Prasad, N.K.; Seiple, I.B.; Cirz, R.T.; Rosenberg, O.S. Leaks in the pipeline: A failure analysis of Gram-negative antibiotic development from 2010 to 2020. Antimicrob. Agents Chemother. 2022, 66, e0005422. [Google Scholar] [CrossRef]
- Voser, T.M.; Campbell, M.D.; Carroll, A.R. How different are marine microbial natural products compared to their terrestrial counterparts? Nat. Prod. Rep. 2022, 39, 7–19. [Google Scholar] [CrossRef]
- Melander, R.J.; Basak, A.K.; Melander, C. Natural products as inspiration for the development of bacterial antibiofilm agents. Nat. Prod. Rep. 2020, 37, 1454–1477. [Google Scholar] [CrossRef] [PubMed]
- Tortorella, E.; Tedesco, P.; Palma Esposito, F.; January, G.G.; Fani, R.; Jaspars, M.; de Pascale, D. Antibiotics from deep-sea microorganisms: Current discoveries and perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; El-Hossary, E.M.; Oelschlaeger, T.A.; Donia, M.S.; Quinn, R.J.; Abdelmohsen, U.R. Potential of marine natural products against drug-resistant bacterial infections. Lancet Infect. Dis. 2019, 19, e237–e245. [Google Scholar] [CrossRef]
- Wiese, J.; Imhoff, J.F. Marine bacteria and fungi as promising source for new antibiotics. Drug Dev. Res. 2019, 80, 24–27. [Google Scholar] [CrossRef] [Green Version]
- Avila, C.; Angulo-Preckler, C. Bioactive compounds from marine heterobranchs. Mar. Drugs 2020, 18, 657. [Google Scholar] [CrossRef]
- Barbosa, F.; Pinto, E.; Kijjoa, A.; Pinto, M.; Sousa, E. Targeting antimicrobial drug resistance with marine natural products. Int. J. Antimicrob. Agents 2020, 56, 106005. [Google Scholar] [CrossRef]
- Bech, P.K.; Lysdal, K.L.; Gram, L.; Bentzon-Tilia, M.; Strube, M.L. Marine sediments hold an untapped potential for novel taxonomic and bioactive bacterial diversity. mSystems 2020, 5, e00782-20. [Google Scholar] [CrossRef] [PubMed]
- Durães, F.; Szemerédi, N.; Kumla, D.; Pinto, M.; Kijjoa, A.; Spengler, G.; Sousa, E. Metabolites from marine-derived fungi as potential antimicrobial adjuvants. Mar. Drugs 2021, 19, 475. [Google Scholar] [CrossRef]
- Nweze, J.A.; Mbaoji, F.N.; Huang, G.; Li, Y.; Yang, L.; Zhang, Y.; Huang, S.; Pan, L.; Yang, D. Antibiotics development and the potentials of marine-derived compounds to stem the tide of multidrug-resistant pathogenic bacteria, fungi, and protozoa. Mar. Drugs 2020, 18, 145. [Google Scholar] [CrossRef] [Green Version]
- Willems, T.; De Mol, M.L.; De Bruycker, A.; De Maeseneire, S.L.; Soetaert, W.K. Alkaloids from marine fungi: Promising antimicrobials. Antibiotics 2020, 9, 340. [Google Scholar] [CrossRef]
- Stincone, P.; Brandelli, A. Marine bacteria as source of antimicrobial compounds. Crit. Rev. Biotechnol. 2020, 40, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.G.M.; Madureira-Carvalho, Á.; Dias-da-Silva, D.; Valentão, P.; Andrade, P.B. Biosynthetic versatility of marine-derived fungi on the delivery of novel antibacterial agents against priority pathogens. Biomed. Pharmacother. 2021, 140, 111756. [Google Scholar] [CrossRef]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine bacterial secondary metabolites: A treasure house for structurally unique and effective antimicrobial compounds. Mar. Drugs 2021, 19, 530. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Maimaitiming, M.; Zhou, Y.; Li, H.; Wang, P.; Liu, Y.; Schäberle, T.F.; Liu, Z.; Wang, C.Y. Discovery of marine natural products as promising antibiotics against Pseudomonas aeruginosa. Mar. Drugs 2022, 20, 192. [Google Scholar] [CrossRef]
- Krishna MS, A.; Mohan, S.; Ashitha, K.T.; Chandramouli, M.; Kumaran, A.; Ningaiah, S.; Babu, K.S.; Somappa, S.B. Marine based natural products: Exploring the recent developments in the identification of antimicrobial agents. Chem. Biodivers. 2022, 19, e202200513. [Google Scholar] [CrossRef]
- Huang, H.; Song, Y.; Li, X.; Wang, X.; Ling, C.; Qin, X.; Zhou, Z.; Li, Q.; Wei, X.; Ju, J. Abyssomicin monomers and dimers from the marine-derived Streptomyces koyangensis SCSIO 5802. J. Nat. Prod. 2018, 81, 1892–1898. [Google Scholar] [CrossRef] [PubMed]
- Sadaka, C.; Ellsworth, E.; Hansen, P.R.; Ewin, R.; Damborg, P.; Watts, J.L. Review on abyssomicins: Inhibitors of the chorismate pathway and folate biosynthesis. Molecules 2018, 23, 1371. [Google Scholar] [CrossRef] [Green Version]
- Braddock, A.A.; Theodorakis, E.A. Marine spirotetronates: Biosynthetic edifices that inspire drug discovery. Mar. Drugs 2019, 17, 232. [Google Scholar] [CrossRef] [Green Version]
- Monjas, L.; Fodran, P.; Kollback, J.; Cassani, C.; Olsson, T.; Genheden, M.; Larsson, D.G.J.; Wallentin, C.J. Synthesis and biological evaluation of truncated derivatives of abyssomicin C as antibacterial agents. Beilstein J. Org. Chem. 2019, 15, 1468–1474. [Google Scholar] [CrossRef] [Green Version]
- Fiedler, H.P. Abyssomicins—A 20-year retrospective view. Mar. Drugs 2021, 19, 299. [Google Scholar] [CrossRef] [PubMed]
- Devine, A.J.; Parnell, A.E.; Back, C.R.; Lees, N.R.; Johns, S.T.; Zulkepli, A.Z.; Barringer, R.; Zorn, K.; Stach, J.E.M.; Crump, M.P.; et al. The role of cytochrome P450 AbyV in the final stages of abyssomicin C biosynthesis. Angew. Chem. Int. Ed. 2023, 62, e202213053. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Schadt, H.S.; Ortel, I.; Süssmuth, R.D. Action of atrop-abyssomicin C as an inhibitor of 4-amino-4-deoxychorismate synthase PabB. Angew. Chem. Int. Ed. 2007, 46, 8284–8286. [Google Scholar] [CrossRef]
- Bihelovic, F.; Karadzic, I.; Matovic, R.; Saicic, R.N. Total synthesis and biological evaluation of (–)-atrop-abyssomicin C. Org. Biomol. Chem. 2013, 11, 5413–5424. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.H.; Nam, S.J.; Locke, J.B.; Kauffman, C.A.; Beatty, D.S.; Paul, L.A.; Fenical, W. Anthracimycin, a potent anthrax antibiotic from a marine-derived actinomycete. Angew. Chem. Int. Ed. 2013, 52, 7822–7824. [Google Scholar] [CrossRef]
- Rodríguez, V.; Martín, J.; Sarmiento-Vizcaíno, A.; de la Cruz, M.; García, L.A.; Blanco, G.; Reyes, F. Anthracimycin B, a potent antibiotic against Gram-positive bacteria isolated from cultures of the deep-sea actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs 2018, 16, 406. [Google Scholar] [CrossRef] [Green Version]
- Alt, S.; Wilkinson, B. Biosynthesis of the novel macrolide antibiotic anthracimycin. ACS Chem. Biol. 2015, 10, 2468–2479. [Google Scholar] [CrossRef]
- Jungmann, K.; Jansen, R.; Gerth, K.; Huch, V.; Krug, D.; Fenical, W.; Müller, R. Two of a kind–the biosynthetic pathways of chlorotonil and anthracimycin. ACS Chem. Biol. 2015, 10, 2480–2490. [Google Scholar] [CrossRef]
- Harunari, E.; Komaki, H.; Igarashi, Y. Biosynthetic origin of anthracimycin: A tricyclic macrolide from Streptomyces sp. J. Antibiot. 2016, 69, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ren, Z.; Chunyu, W.X.; Li, G.D.; Chen, X.; Zhang, Z.T.; Sun, H.B.; Wang, M.; Xie, T.P.; Wang, M. Exploration of diverse secondary metabolites from Streptomyces sp. YINM00001, using genome mining and one strain many compounds approach. Front. Microbiol. 2022, 13, 831174. [Google Scholar] [CrossRef]
- Hensler, M.E.; Jang, K.H.; Thienphrapa, W.; Vuong, L.; Tran, D.N.; Soubih, E.; Lin, L.; Haste, N.M.; Cunningham, M.L.; Kwan, B.P.; et al. Anthracimycin activity against contemporary methicillin-resistant Staphylococcus aureus. J. Antibiot. 2014, 67, 549–553. [Google Scholar] [CrossRef] [Green Version]
- Hofer, W.; Oueis, E.; Fayad, A.A.; Deschner, F.; Andreas, A.; de Carvalho, L.P.; Hüttel, S.; Bernecker, S.; Pätzold, L.; Morgenstern, B.; et al. Regio- and stereoselective epoxidation and acidic epoxide opening of antibacterial and antiplasmodial chlorotonils yield highly potent derivatives. Angew. Chem. Int. Ed. 2022, 61, e202202816. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.C.; Lim, A.L.; Pond, C.D.; Craft, M.; Čavužić, M.; Waldrop, G.L.; Schmidt, E.W.; Barrows, L.R. Pyrrolocin C and equisetin inhibit bacterial acetyl-CoA carboxylase. PLoS ONE 2020, 15, e0233485. [Google Scholar] [CrossRef]
- Fujiyama, K.; Kato, N.; Re, S.; Kinugasa, K.; Watanabe, K.; Takita, R.; Nogawa, T.; Hino, T.; Osada, H.; Sugita, Y.; et al. Molecular basis for two stereoselective Diels-Alderases that produce decalin skeletons. Angew. Chem. Int Ed. 2021, 60, 22401–22410. [Google Scholar] [CrossRef]
- Chi, C.; Wang, Z.; Liu, T.; Zhang, Z.; Zhou, H.; Li, A.; Jin, H.; Jia, H.; Yin, F.; Yang, D. Crystal structures of Fsa2 and Phm7 catalyzing [4 + 2] cycloaddition reactions with reverse stereoselectivities in equisetin and phomasetin biosynthesis. ACS Omega 2021, 6, 12913–12922. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, D.; Zhang, Q.; Guo, P.; Ding, S.; Shen, J.; Zhu, K.; Lin, W. A marine antibiotic kills multidrug-resistant bacteria without detectable high-level resistance. ACS Infect. Dis. 2021, 7, 884–893. [Google Scholar] [CrossRef]
- Tian, J.; Chen, S.; Liu, F.; Zhu, Q.; Shen, J.; Lin, W.; Zhu, K. Equisetin targets intracellular Staphylococcus aureus through a host acting strategy. Mar. Drugs 2022, 20, 656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, S.; Liu, X.; Lin, W.; Zhu, K. Equisetin restores colistin sensitivity against multi-drug resistant Gram-negative bacteria. Antibiotics 2021, 10, 1263. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kim, H.J.; Kim, S.; Park, S.Y.; Kim, H.; Jeong, S.; Lee, S.J.; Lee, M.S. Enhanced large-scale production of Hahella chejuensis-derived prodigiosin and evaluation of Its bioactivity. J. Microbiol. Biotechnol. 2021, 31, 1624–1631. [Google Scholar] [CrossRef]
- Sakai-Kawada, F.E.; Ip, C.G.; Hagiwara, K.A.; Awaya, J.D. Biosynthesis and bioactivity of prodiginine analogs in marine bacteria, Pseudoalteromonas: A mini review. Front. Microbiol. 2019, 10, 1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattingly, A.E.; Cox, K.E.; Smith, R.; Melander, R.J.; Ernst, R.K.; Melander, C. Screening an established natural product library identifies secondary metabolites that potentiate conventional antibiotics. ACS Infect. Dis. 2020, 6, 2629–2640. [Google Scholar] [CrossRef]
- He, S.; Li, P.; Wang, J.; Zhang, Y.; Lu, H.; Shi, L.; Huang, T.; Zhang, W.; Ding, L.; He, S. Discovery of new secondary metabolites from marine bacteria Hahella based on an omics strategy. Mar. Drugs 2022, 20, 269. [Google Scholar] [CrossRef] [PubMed]
- Siwawannapong, K.; Nemeth, A.M.; Melander, R.J.; Rong, J.; Davis, J.R.; Taniguchi, M.; Carpenter, M.E.; Lindsey, J.S.; Melander, C. Simple dipyrrin analogues of prodigiosin for use as colistin adjuvants. ChemMedChem 2022, 17, e202200286. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Waters, A.L.; Sims, J.W.; Fullmer, A.; Ellison, S.; Hamann, M.T. Complex marine natural products as potential epigenetic and production regulators of antibiotics from a marine Pseudomonas aeruginosa. Microb. Ecol. 2013, 65, 1068–1075. [Google Scholar] [CrossRef] [Green Version]
- Pech-Puch, D.; Pérez-Povedano, M.; Martinez-Guitian, M.; Lasarte-Monterrubio, C.; Vázquez-Ucha, J.C.; Bou, G.; Rodríguez, J.; Beceiro, A.; Jimenez, C. In vitro and in vivo assessment of the efficacy of bromoageliferin, an alkaloid isolated from the sponge Agelas dilatata, against Pseudomonas aeruginosa. Mar. Drugs 2020, 18, 326. [Google Scholar] [CrossRef]
- Freire, V.F.; Gubiani, J.R.; Spencer, T.M.; Hajdu, E.; Ferreira, A.G.; Ferreira, D.A.S.; de Castro Levatti, E.V.; Burdette, J.E.; Camargo, C.H.; Tempone, A.G.; et al. Feature-based molecular networking discovery of bromopyrrole alkaloids from the marine sponge Agelas dispar. J. Nat. Prod. 2022, 85, 1340–1350. [Google Scholar] [CrossRef]
- Bernan, V.S.; Roll, D.M.; Ireland, C.M.; Greenstein, M.; Maiese, W.M.; Steinberg, D.A. A study on the mechanism of action of sceptrin, an antimicrobial agent isolated from the South Pacific sponge Agelas mauritiana. J. Antimicrob. Chemother. 1993, 32, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.D.; Lear, M.J.; La Clair, J.J. Identification of the binding of sceptrin to MreB via a bidirectional affinity protocol. J. Am. Chem. Soc. 2008, 130, 7256–7258. [Google Scholar] [CrossRef]
- Keffer, J.L.; Huecas, S.; Hammill, J.T.; Wipf, P.; Andreu, J.M.; Bewley, C.A. Chrysophaentins are competitive inhibitors of FtsZ and inhibit Z-ring formation in live bacteria. Bioorg. Med. Chem. 2013, 21, 5673–5678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davison, J.R.; Bewley, C.A. Antimicrobial chrysophaentin analogs identified from laboratory cultures of the marine microalga Chrysophaeum taylorii. J. Nat. Prod. 2019, 82, 148–153. [Google Scholar] [CrossRef]
- Fullenkamp, C.R.; Hsu, Y.P.; Quardokus, E.M.; Zhao, G.; Bewley, C.A.; VanNieuwenhze, M.; Sulikowski, G.A. Synthesis of 9-dechlorochrysophaentin A enables studies revealing bacterial cell wall biosynthesis inhibition phenotype in B. subtilis. J. Am. Chem. Soc. 2020, 142, 16161–16166. [Google Scholar] [CrossRef]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltz, R.H. Genome mining for drug discovery: Cyclic lipopeptides related to daptomycin. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab020. [Google Scholar] [CrossRef]
- Wood, T.M.; Zeronian, M.R.; Buijs, N.; Bertheussen, K.; Abedian, H.K.; Johnson, A.V.; Pearce, N.M.; Lutz, M.; Kemmink, J.; Seirsma, T.; et al. Mechanistic insights into the C55-P targeting lipopeptide antibiotics revealed by structure-activity studies and high-resolution crystal structures. Chem. Sci. 2022, 13, 2985–2991. [Google Scholar] [CrossRef]
- Taylor, S.D. A decade of research on daptomycin. Synlett 2022, 33, 1695–1706. [Google Scholar] [CrossRef]
- Tomoda, H. New approaches to drug discovery for combating MRSA. Chem. Pharm. Bull. 2016, 64, 104–111. [Google Scholar] [CrossRef]
- Ikeda, H.; Shin-Ya, K.; Nagamitsu, T.; Tomoda, H. Biosynthesis of mercapturic acid derivative of the labdane-type diterpene, cyslabdan that potentiates imipenem activity against methicillin-resistant Staphylococcus aureus: Cyslabdan is generated by mycothiol-mediated xenobiotic detoxification. J. Ind. Microbiol. Biotechnol. 2016, 43, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Trampe, S.M.; Ikeda, H.; Vinuesa, P.; Macías-Rubalcava, M.L.; Esquivel, B.; Centeno-Leija, S.; Tapia-Cabrera, S.M.; Mora-Herrera, S.I.; Ruiz-Villafán, B.; Rodríguez-Sanoja, R. Production of distinct labdane-type diterpenoids using a novel cryptic labdane-like cluster from Streptomyces thermocarboxydus K155. Appl. Microbiol. Biotechnol. 2020, 104, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Koyama, N.; Tokura, Y.; Münch, D.; Sahl, H.G.; Schneider, T.; Shibagaki, Y.; Ikeda, H.; Tomoda, H. The nonantibiotic small molecule cyslabdan enhances the potency of β-lactams against MRSA by inhibiting pentaglycine interpeptide bridge synthesis. PLoS ONE 2012, 7, e48981. [Google Scholar] [CrossRef] [Green Version]
- Fukumoto, A.; Kim, Y.P.; Hanaki, H.; Shiomi, K.; Tomoda, H.; Omura, S. Cyslabdan, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Streptomyces sp. K04-0144. II. Biological activities. J. Antibiot. 2008, 61, 7–10. [Google Scholar] [CrossRef] [Green Version]
- Shanthi, J.; Senthil, A.; Gopikrishnan, V.; Balagurunathan, R. Characterization of a potential β-lactamase inhibitory metabolite from a marine Streptomyces sp. PM49 active against multidrug-resistant pathogens. Appl. Biochem. Biotechnol. 2015, 175, 3696–3708. [Google Scholar] [CrossRef] [PubMed]
- Ohtawa, M.; Hishinuma, Y.; Takagi, E.; Yamada, T.; Ito, F.; Arima, S.; Uchida, R.; Kim, Y.P.; Ōmura, S.; Tomoda, H.; et al. Synthesis and structural revision of cyslabdan. Chem. Pharm. Bull. 2016, 64, 1370–1377. [Google Scholar] [CrossRef] [Green Version]
- Mills, J.J.; Robinson, K.R.; Zehnder, T.E.; Pierce, J.G. Synthesis and biological evaluation of the antimicrobial natural product lipoxazolidinone A. Angew. Chem. Int. Ed. 2018, 57, 8682–8686. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Pena, M.A.; Massaro, N.P.; Lin, Y.C.; Pierce, J.G. Leveraging marine natural products as a platform to tackle bacterial resistance and persistence. Acc. Chem. Res. 2021, 54, 1866–1877. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.R.; Mills, J.J.; Pierce, J.G. Expanded structure-activity studies of lipoxazolidinone antibiotics. ACS Med. Chem. Lett. 2019, 10, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Mary, T.R.J.; Kannan, R.R.; Iniyan, A.M.; Ranjith, W.A.C.; Nandhagopal, S.; Vishwakarma, V.; Vincent, S.G.P. β-Lactamase inhibitory potential of kalafungin from marine Streptomyces in Staphylococcus aureus infected zebrafish. Microbiol. Res. 2021, 244, 126666. [Google Scholar] [CrossRef]
- Jeong, B.G.; Na, J.H.; Bae, D.W.; Park, S.B.; Lee, H.S.; Cha, S.S. Crystal structure of AmpC BER and molecular docking lead to the discovery of broad inhibition activities of halisulfates against β-lactamases. Comput. Struct. Biotechnol. J. 2021, 19, 145–152. [Google Scholar] [CrossRef]
- Liu, S.; Su, M.; Song, S.J.; Jung, J.H. Marine-derived Penicillium species as producers of cytotoxic metabolites. Mar. Drugs 2017, 15, 329. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.F.; Lee, S.K.; Price, J.; Jack, R.W.; Turner, G.; Kong, R.Y. Cloning and expression analysis of the pcbAB-pcbC β-lactam genes in the marine fungus Kallichroma tethys. Appl. Environ. Microbiol. 2003, 69, 1308–1314. [Google Scholar] [CrossRef] [Green Version]
- Hamed, R.B.; Gomez-Castellanos, J.R.; Henry, L.; Ducho, C.; McDonough, M.A.; Schofield, C.J. The enzymes of β-lactam biosynthesis. Nat. Prod. Rep. 2013, 30, 21–107. [Google Scholar] [CrossRef]
- Townsend, C.A. Convergent biosynthetic pathways to β-lactam antibiotics. Curr. Opin. Chem. Biol. 2016, 35, 97–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabe, P.; Kamps, J.J.A.G.; Schofield, C.J.; Lohans, C.T. Roles of 2-oxoglutarate oxygenases and isopenicillin N synthase in β-lactam biosynthesis. Nat. Prod. Rep. 2018, 35, 735–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scannell, J.P.; Pruess, D.L.; Blount, J.F.; Ax, H.A.; Kellett, M.; Weiss, F.; Demny, T.C.; Williams, T.H.; Stempel, A. Antimetabolites produced by microorganisms. XII. (S)-Alanyl-3-[α(S)-chloro-3-(S)-hydroxy-2-oxo-3-azetidinylmethyl]-(S)-alanine, a new β-lactam containing natural product. J. Antibiot. 1975, 28, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Manning, M.E.; Danson, E.J.; Calderone, C.T. Functional chararacterization of the enzymes TabB and TabD involved in tabtoxin biosynthesis by Pseudomonas syringae. Biochem. Biophys. Res. Commun. 2018, 496, 212–217. [Google Scholar] [CrossRef]
- Lyu, J.; Ushimaru, R.; Abe, I. Characterization of enzymes catalyzing the initial steps of the β-lactam tabtoxin biosynthesis. Org. Lett. 2022, 24, 3337–3341. [Google Scholar] [CrossRef] [PubMed]
- Hart, K.M.; Reck, M.; Bowman, G.R.; Wencewicz, T.A. Tabtoxinine-β-lactam is a “stealth” β-lactam antibiotic that evades β-lactamase-mediated antibiotic resistance. Med. Chem. Commun. 2016, 7, 118–127. [Google Scholar] [CrossRef]
- Patrick, G.J.; Fang, L.; Schaefer, J.; Singh, S.; Bowman, G.R.; Wencewicz, T.A. Mechanistic basis for ATP-dependent inhibition of glutamine synthetase by tabtoxinine-β-lactam. Biochemistry 2018, 57, 117–135. [Google Scholar] [CrossRef]
- Anthoni, U.; Bock, K.; Chevolot, L.; Larsen, C.; Nielsen, P.H.; Christophersen, C. Marine alkaloids. 13. Chartellamide A and B, halogenated β-lactam indole-imidazole alkaloids from the marine bryozoan Chartella papyracea. J. Org. Chem. 1987, 52, 5638–5639. [Google Scholar] [CrossRef]
- Avilés, E.; Rodríguez, A.D. Monamphilectine A, a potent antimalarial β-lactam from marine sponge Hymeniacidon sp: Isolation, structure, semisynthesis, and bioactivity. Org. Lett. 2010, 12, 5290–5293. [Google Scholar] [CrossRef] [Green Version]
- Avilés, E.; Prudhomme, J.; Le Roch, K.G.; Rodríguez, A.D. Structures, semisyntheses, and absolute configurations of the antiplasmodial α-substituted β-lactam monamphilectines B and C from the sponge Svenzea flava. Tetrahedron 2015, 71, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duff, J.P.; AbuOun, M.; Bexton, S.; Rogers, J.; Turton, J.; Woodford, N.; Irvine, R.; Anjum, M.; Teale, C. Resistance to carbapenems and other antibiotics in Klebsiella pneumoniae found in seals indicates anthropogenic pollution. Vet. Rec. 2020, 187, 154. [Google Scholar] [CrossRef] [PubMed]
- Hatosy, S.M.; Martiny, A.C. The ocean as a global reservoir of antibiotic resistance genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef] [Green Version]
- Elbehery, A.H.; Leak, D.J.; Siam, R. Novel thermostable antibiotic resistance enzymes from the Atlantis II Deep Red Sea brine pool. Microb. Biotechnol. 2017, 10, 189–202. [Google Scholar] [CrossRef]
- Tan, L.; Li, L.; Ashbolt, N.; Wang, X.; Cui, Y.; Zhu, X.; Xu, Y.; Yang, Y.; Mao, D.; Luo, Y. Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci. Total Environ. 2018, 621, 1176–1184. [Google Scholar] [CrossRef]
- Blanco-Picazo, P.; Roscales, G.; Toribio-Avedillo, D.; Gómez-Gómez, C.; Avila, C.; Ballesté, E.; Muniesa, M.; Rodríguez-Rubio, L. Antibiotic resistance genes in phage particles from antarctic and mediterranean seawater ecosystems. Microorganisms 2020, 8, 1293. [Google Scholar] [CrossRef]
- Cuadrat, R.R.C.; Sorokina, M.; Andrade, B.G.; Goris, T.; Dávila, A.M.R. Global ocean resistome revealed: Exploring antibiotic resistance gene abundance and distribution in TARA Oceans samples. Gigascience 2020, 9, giaa046. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Huang, X.; Zhang, G.; Yuan, L.; Shen, E.; Zhang, L.; Zhang, X.H.; Zhang, T.; Tao, L.; Ju, F. Distinctive signatures of pathogenic and antibiotic resistant potentials in the hadal microbiome. Environ. Microbiome 2022, 17, 19. [Google Scholar] [CrossRef]
- Perry, J.; Waglechner, N.; Wright, G. The prehistory of antibiotic resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025197. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.D. Environmental and clinical antibiotic resistomes, same only different. Curr. Opin. Microbiol. 2019, 51, 57–63. [Google Scholar] [CrossRef]
- Massova, I.; Mobashery, S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob. Agents Chemother. 1998, 42, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Modi, T.; Risso, V.A.; Martinez-Rodriguez, S.; Gavira, J.A.; Mebrat, M.D.; Van Horn, W.D.; Sanchez-Ruiz, J.M.; Ozkan, S.B. Hinge-shift mechanism as a protein design principle for the evolution of β-lactamases from substrate promiscuity to specificity. Nat. Commun. 2021, 12, 1852. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Yoo, W.; Kim, Y.O.; Kim, K.K.; Kim, T.D. Molecular characterization of a novel family VIII esterase with β-lactamase activity (PsEstA) from Paenibacillus sp. Biomolecules 2019, 9, 786. [Google Scholar] [CrossRef] [Green Version]
- Ryu, B.H.; Ngo, T.D.; Yoo, W.; Lee, S.; Kim, B.Y.; Lee, E.; Kim, K.K.; Kim, T.D. Biochemical and structural analysis of a novel esterase from Caulobacter crescentus related to penicillin-rinding protein (PBP). Sci. Rep. 2016, 6, 37978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Lin, X.; Xu, C.; Shen, Y.; Wang, S.-P.; Liao, H.; Li, L.; Deng, H.; Lin, H.-W. Investigation of penicillin binding protein (PBP)-like peptide cyclase and hydrolase in surugamide non-ribosomal peptide biosynthesis. Cell Chem. Biol. 2019, 26, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Kobayashi, M.; Kuranaga, T.; Takada, K.; Ikeda, H.; Matsunaga, S.; Wakimoto, T. SurE is a trans-acting thioesterase cyclizing two distinct non-ribosomal peptides. Org. Biomol. Chem. 2019, 17, 1058–1061. [Google Scholar] [CrossRef]
- Cea-Rama, I.; Coscolín, C.; Gonzalez-Alfonso, J.L.; Raj, J.; Vasiljević, M.; Plou, F.J.; Ferrer, M.; Sanz-Aparicio, J. Crystal structure of a family VIII β-lactamase fold hydrolase reveals the molecular mechanism for its broad substrate scope. FEBS J. 2022, 289, 6714–6730. [Google Scholar] [CrossRef]
- Jeon, J.H.; Lee, H.S.; Lee, J.H.; Koo, B.S.; Lee, C.M.; Lee, S.H.; Kang, S.G.; Lee, J.H. A novel family VIII carboxylesterase hydrolysing third- and fourth-generation cephalosporins. Springerplus 2016, 5, 525. [Google Scholar] [CrossRef] [Green Version]
- Schuster, M.; Sexton, D.J.; Diggle, S.P.; Greenberg, E.P. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu. Rev. Microbiol. 2013, 67, 43–63. [Google Scholar] [CrossRef]
- Xavier, K.B.; Bassler, B.L. LuxS quorum sensing: More than just a numbers game. Curr. Opin. Microbiol. 2003, 6, 191–197. [Google Scholar] [CrossRef]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [Green Version]
- Sharifzadeh, S.; Brown, N.W.; Shirley, J.D.; Bruce, K.E.; Winkler, M.E.; Carlson, E.E. Chemical tools for selective activity profiling of bacterial penicillin-binding proteins. Methods Enzymol. 2020, 638, 27–55. [Google Scholar] [CrossRef]
- Brown, N.W.; Shirley, J.; Marshall, A.; Carlson, E. Comparison of bioorthogonal β-lactone activity-based probes for selective labeling of penicillin-binding proteins. ChemBioChem 2021, 22, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Flanders, P.L.; Contreras-Martel, C.; Brown, N.W.; Shirley, J.D.; Martins, A.; Nauta, K.N.; Dessen, A.; Carlson, E.E.; Ambrose, E.A. Combined structural analysis and molecular dynamics reveal penicillin-binding protein inhibition mode with β-lactones. ACS Chem. Biol. 2022, 17, 3110–3120. [Google Scholar] [CrossRef] [PubMed]
- Aertker, K.M.J.; Chan, H.T.H.; Lohans, C.T.; Schofield, C.J. Analysis of β-lactone formation by clinically observed carbapenemases informs on a novel antibiotic resistance mechanism. J. Biol. Chem. 2020, 295, 16604–16613. [Google Scholar] [CrossRef] [PubMed]
- Lohans, C.T.; van Groesen, E.; Kumar, K.; Tooke, C.L.; Spencer, J.; Paton, R.S.; Brem, J.; Schofield, C. A new mechanism for β-lactamases: Class D enzymes degrade 1β-methyl carbapenems via lactone formation. Angew. Chem. Int. Ed. 2018, 57, 1282–1285. [Google Scholar] [CrossRef] [Green Version]
- Macheboeuf, P.; Fischer, D.S.; Brown Jr., T.; Zervosen, A.; Luxen, A.; Joris, B.; Dessen, A.; Schofield, C.J. Structural and mechanistic basis of penicillin-binding protein inhibition by lactivicins. Nat. Chem. Biol. 2007, 3, 565–569. [Google Scholar] [CrossRef]
- Brown Jr., T.; Charlier, P.; Herman, R.; Schofield, C.J.; Sauvage, E. Structural basis for the interaction of lactivicins with serine β-lactamases. J. Med. Chem. 2010, 53, 5890–5894. [Google Scholar] [CrossRef] [PubMed]
- Kluge, A.F.; Petter, R.C. Acylating drugs: Redesigning natural covalent inhibitors. Curr. Opin. Chem. Biol. 2010, 14, 421–427. [Google Scholar] [CrossRef]
- Böttcher, T.; Sieber, S.A. β-Lactams and β-lactones as activity-based probes in chemical biology. Med. Chem. Commun. 2012, 3, 408–417. [Google Scholar] [CrossRef]
- Wiedemann, E.N.; Mandl, F.A.; Blank, I.D.; Ochsenfeld, C.; Ofial, A.R.; Sieber, S.A. Kinetic and theoretical studies of β-lactone reactivity—A quantitative scale for biological application. ChemPlusChem 2015, 80, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Mazur, M.; Maslowiec, D. Antimicrobial activity of lactones. Antibiotics 2022, 11, 1327. [Google Scholar] [CrossRef]
- Liu, J.; Fu, K.; Wu, C.; Qin, K.; Li, F.; Zhou, L. “In-Group” communication in marine Vibrio: A review of N-acyl homoserine lactones-driven quorum sensing. Front. Cell. Infect. Microbiol. 2018, 8, 139. [Google Scholar] [CrossRef]
- Majik, M.S.; Gawas, U.B.; Mandrekar, V.K. Next generation quorum sensing inhibitors: Accounts on structure activity relationship studies and biological activities. Bioorg. Med. Chem. 2020, 28, 115728. [Google Scholar] [CrossRef]
- Polaske, T.J.; Gahan, C.G.; Nyffeler, K.E.; Lynn, D.M.; Blackwell, H.E. Identification of small molecules that strongly inhibit bacterial quorum sensing using a high-throughput lipid vesicle lysis assay. Cell Chem. Biol. 2022, 29, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Simões, M. Quorum sensing inhibition by marine bacteria. Mar. Drugs 2019, 17, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, S.-F.; Chao, Y.-F.; Lin, J.-W. Identification and characteristic analysis of the ampC gene encoding β-lactamase from Vibrio fischeri. Biochem. Biophys. Res. Commun. 2004, 314, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Toth, M.; Smith, C.; Frase, H.; Mobashery, S.; Vakulenko, S. An antibiotic-resistance enzyme from a deep-sea bacterium. J. Am. Chem. Soc. 2010, 132, 816–823. [Google Scholar] [CrossRef] [Green Version]
- Pietra, F. On 3LEZ, a deep-sea halophilic protein with in vitro class-A β-lactamase activity: Molecular-dynamics, docking, and reactivity simulations. Chem. Biodivers. 2012, 9, 2659–2684. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.W.; Cheng, H.; Huo, Y.Y.; Xu, L.; Wu, Y.H.; Liu, W.H.; Tao, F.F.; Cui, X.J.; Zheng, B.W. Biochemical and genetic characterization of a novel metallo-β-lactamase from marine bacterium Erythrobacter litoralis HTCC 2594. Sci. Rep. 2018, 8, 803. [Google Scholar] [CrossRef] [Green Version]
- Kieffer, N.; Guzmán-Puche, J.; Poirel, L.; Kang, H.J.; Jeon, C.O.; Nordmann, P. ZHO-1, an intrinsic MBL from the environmental Gram-negative species Zhongshania aliphaticivorans. J. Antimicrob. Chemother. 2019, 74, 1568–1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selleck, C.; Pedroso, M.M.; Wilson, L.; Krco, S.; Knaven, E.G.; Miraula, M.; Mitić, N.; Larrabee, J.A.; Brück, T.; Clark, A. Structure and mechanism of potent bifunctional β-lactam- and homoserine lactone-degrading enzymes from marine microorganisms. Sci. Rep. 2020, 10, 12882. [Google Scholar] [CrossRef]
- Gersch, M.; Kreuzer, J.; Sieber, S.A. Electrophilic natural products and their biological targets. Nat. Prod. Rep. 2012, 29, 659–682. [Google Scholar] [CrossRef] [PubMed]
- Garner, A.L.; Yu, J.; Struss, A.K.; Kaufmann, G.F.; Kravchenko, V.V.; Janda, K.D. Immunomodulation and the quorum sensing molecule 3-oxo-C12-homoserine lactone: The importance of chemical scaffolding for probe development. Chem. Commun. 2013, 49, 1515–1517. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Lorenz, N.; Jung, K.; Sieber, S.A. Mechanistic analysis of aliphatic β-lactones in Vibrio harveyi reveals a quorum sensing independent mode of action. Chem. Commun. 2016, 52, 11971–11974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottcher, T.; Sieber, S.A. β-Lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 2008, 130, 14400–14401. [Google Scholar] [CrossRef]
- Gersch, M.; Gut, F.; Korotkov, V.S.; Lehmann, J.; Bottcher, T.; Rusch, M.; Hedberg, C.; Waldmann, H.; Klebe, G.; Sieber, S.A. The mechanism of caseinolytic protease (ClpP) inhibition. Angew. Chem. Int. Ed. 2013, 52, 3009–3014. [Google Scholar] [CrossRef]
- Krysiak, J.; Stahl, M.; Vomacka, J.; Fetzer, C.; Lakemeyer, M.; Fux, A.; Sieber, S.A. Quantitative map of β-lactone-induced virulence regulation. J. Proteome Res. 2017, 16, 1180–1192. [Google Scholar] [CrossRef]
- Delago, A.; Gregor, R.; Dubinsky, L.; Dandela, R.; Hendler, A.; Krief, P.; Rayo, J.; Aharoni, A.; Meijler, M.M. A bacterial quorum sensing molecule elicits a general stress response in Saccharomyces cerevisiae. Front. Microbiol. 2021, 12, 632658. [Google Scholar] [CrossRef]
- Rayo, J.; Gregor, R.; Jacob, N.T.; Dandela, R.; Dubinsky, L.; Yashkin, A.; Aranovich, A.; Thangaraj, M.; Ernst, O.; Barash, E.; et al. Immunoediting role for major vault protein in apoptotic signaling induced by bacterial N-acyl homoserine lactones. Proc. Natl. Acad. Sci. USA 2021, 118, e2012529118. [Google Scholar] [CrossRef]
- Kreitler, D.F.; Gemmell, E.M.; Schaffer, J.E.; Wencewicz, T.A.; Gulick, A.M. The structural basis of N-acyl-α-amino-β-lactone formation catalyzed by a nonribosomal peptide synthetase. Nat. Commun. 2019, 10, 3432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, T.A.; Batey, S.F.D.; Wiencek, P.; Chandra, G.; Alt, S.; Francklyn, C.S.; Wilkinson, B. Immunity-guided identification of threonyl-tRNA synthetase as the molecular target of obafluorin, a β-lactone antibiotic. ACS Chem. Biol. 2019, 14, 2663–2671. [Google Scholar] [CrossRef] [PubMed]
- Travin, D.Y.; Severinov, K.; Dubiley, S. Natural Trojan horse inhibitors of aminoacyl-tRNA synthetases. RSC Chem. Biol. 2021, 2, 468–485. [Google Scholar] [CrossRef]
- Schaffer, J.E.; Reck, M.R.; Prasad, N.K.; Wencewicz, T.A. β-Lactone formation during product release from a nonribosomal peptide synthetase. Nat. Chem. Biol. 2017, 13, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Lakemeyer, M.; Zhao, W.; Mandl, F.A.; Hammann, P.; Sieber, S.A. Thinking outside the box–novel antibacterials to tackle the resistance crisis. Angew. Chem. Int. Ed. 2018, 57, 14440–14475. [Google Scholar] [CrossRef]
- Cook, M.A.; Wright, G.D. The past, present, and future of antibiotics. Sci. Transl. Med. 2022, 14, eabo7793. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fisher, J.F.; Mobashery, S. β-Lactams from the Ocean. Mar. Drugs 2023, 21, 86. https://doi.org/10.3390/md21020086
Fisher JF, Mobashery S. β-Lactams from the Ocean. Marine Drugs. 2023; 21(2):86. https://doi.org/10.3390/md21020086
Chicago/Turabian StyleFisher, Jed F., and Shahriar Mobashery. 2023. "β-Lactams from the Ocean" Marine Drugs 21, no. 2: 86. https://doi.org/10.3390/md21020086
APA StyleFisher, J. F., & Mobashery, S. (2023). β-Lactams from the Ocean. Marine Drugs, 21(2), 86. https://doi.org/10.3390/md21020086