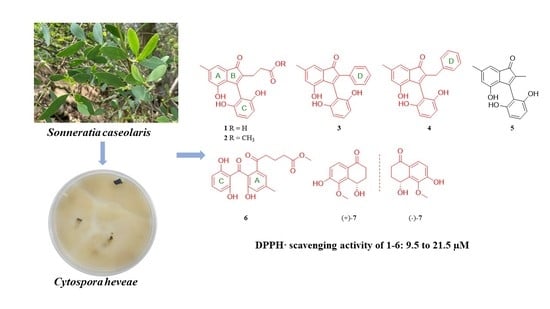
Antioxidative Indenone and Benzophenone Derivatives from the Mangrove-Derived Fungus Cytospora heveae NSHSJ-2
Abstract
:1. Introduction
2. Results
2.1. Structure Elucidation
2.2. Biological Evaluation
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction and Isolation
3.4. Biological Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prasher, P.; Sharma, M. Medicinal chemistry of indane and its analogues: A mini review. ChemistrySelect 2021, 6, 2658–2677. [Google Scholar] [CrossRef]
- Nigam, R.; Babu, K.R.; Ghosh, T.; Kumari, B.; Akula, D.; Rath, S.N.; Das, P.; Anindya, R.; Khan, F.A. Indenone derivatives as inhibitor of human DNA dealkylation repair enzyme AlkBH3. Bioorgan. Med. Chem. 2018, 26, 4100–4112. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.D.; Chang, J.; Qin, B.Y.; Zhong, C.; Chu, Z.B.; Huang, J.; Zhou, W.J.; Sun, X. Synthesis, estrogenic activity, and anti-osteoporosis effects in ovariectomized rats of resveratrol oligomer derivatives. Eur. J. Med. Chem. 2015, 102, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Shin, M.S.; Jung, S.H.; Kang, S.K.; Kim, K.R.; Rhee, S.D.; Jung, W.H.; Yang, S.D.; Kim, S.J.; Woo, J.R.; et al. Indenone Derivatives: A Novel Template for Peroxisome Proliferator-Activated Receptor γ (PPARγ) Agonists. J. Med. Chem. 2006, 49, 4781–4784. [Google Scholar] [CrossRef]
- Liu, J.; Liu, L.; Zheng, L.; Feng, K.W.; Wang, H.T.; Xu, J.P.; Zhou, Z.Z. Discovery of novel 2,3-dihydro-1H-inden-1-ones as dual PDE4/AChE inhibitors with more potency against neuroinflammation for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2022, 238, 114503. [Google Scholar] [CrossRef] [PubMed]
- Ernst-Russell, M.A.; Chai, C.L.L.; Wardlaw, J.H.; Elix, J.A. Euplectin and Coneuplectin, New Naphthopyrones from the Lichen Flavoparmelia euplecta. J. Nat. Prod. 2000, 63, 129–131. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Ru, T.; Navarro-Vázquez, A.; Lindemann, P.; Nazaré, M.; Li, X.W.; Guo, Y.W.; Sun, H. Weizhouochrones: Gorgonian-Derived Symmetric Dimers and Their Structure Elucidation Using Anisotropic NMR Combined with DP4+ Probability and CASE-3D. J. Nat. Prod. 2022, 85, 1730–1737. [Google Scholar] [CrossRef]
- Du, Y.E.; Byun, W.S.; Lee, S.B.; Hwang, S.; Shin, Y.H.; Shin, B.; Jang, Y.J.; Hong, S.; Shin, J.; Lee, S.K.; et al. Formicins, N-Acetylcysteamine-Bearing Indenone Thioesters from a Wood Ant-Associated Bacterium. Org. Lett. 2020, 22, 5337–5341. [Google Scholar] [CrossRef]
- Luo, H.F.; Zhang, L.P.; Hu, C.Q. Five novel oligostilbenes from the roots of Caragana sinica. Tetrahedron 2001, 57, 4849–4854. [Google Scholar] [CrossRef]
- Sema, D.K.; Lannang, A.M.; Tatsimo, S.J.N.; Rehman, M.; Yousuf, S.; Zoufou, D.; Iqbal, U.; Wansi, J.D.; Sewald, N.; Choudhary, M.I. New indane and naphthalene derivatives from the rhizomes of Kniphofia reflexa Hutchinson ex Codd. Phytochem. Lett. 2018, 26, 78–82. [Google Scholar] [CrossRef]
- Jaki, B.; Heilmann, J.; Sticher, O. New Antibacterial Metabolites from the Cyanobacterium Nostoc commune (EAWAG 122b). J. Nat. Prod. 2000, 63, 1283–1285. [Google Scholar] [CrossRef]
- Kim, H.; Yang, I.; Ryu, S.Y.; Won, D.H.; Giri, A.G.; Wang, W.H.; Choi, H.; Chin, J.; Hahn, D.; Kim, E.; et al. Acredinones A and B, Voltage-Dependent Potassium Channel Inhibitors from the Sponge-Derived Fungus Acremonium sp. F9A015. J. Nat. Prod. 2015, 78, 363–367. [Google Scholar] [CrossRef]
- Liu, Z.M.; Qiu, P.; Li, J.; Chen, G.Y.; Chen, Y.; Liu, H.J.; She, Z.G. Anti-inflammatory polyketides from the mangrove-derived fungus Ascomycota sp. SK2YWS-L. Tetrahedron 2018, 74, 746–751. [Google Scholar] [CrossRef]
- Tan, C.B.; Liu, Z.M.; Chen, S.H.; Huang, X.S.; Cui, H.; Long, Y.H.; Lu, Y.J.; She, Z.G. Antioxidative Polyketones from the Mangrove-Derived Fungus Ascomycota sp. SK2YWS-L. Sci. Rep. 2016, 6, 36609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, C.; Liu, X.H.; Chang, J.; Yu, J.M.; Sun, X. Inhibitory effect of resveratrol dimerized derivatives on nitric oxide production in lipopolysaccharide-induced RAW 264.7 cells. Bioorg. Med. Chem. Lett. 2013, 23, 4413–4418. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Cai, R.L.; Liu, Z.M.; Cui, H.; She, Z.G. Secondary metabolites from mangrove-associated fungi: Source, chemistry and bioactivities. Nat. Prod. Rep. 2022, 39, 560–595. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Bioactive natural products derived from mangrove-associated microbes. RSC Adv. 2015, 5, 841–892. [Google Scholar] [CrossRef]
- Tan, Q.; Yang, W.C.; Zhu, G.; Chen, T.; Wu, J.; Zhu, Y.J.; Wang, B.; Yuan, J.; She, Z.G. A Pair of Chromone Epimers and an Acetophenone Glucoside from the Mangrove Endophytic Fungus Mycosphaerella sp. L3A1. Chem. Biodivers. 2022, 19, e202200998. [Google Scholar] [CrossRef]
- Chen, T.; Yang, W.C.; Li, T.B.; Yin, Y.H.; Liu, Y.F.; Wang, B.; She, Z.G. Hemiacetalmeroterpenoids A–C and Astellolide Q with Antimicrobial Activity from the Marine-Derived Fungus Penicillium sp. N-5. Mar. Drugs 2022, 20, 514. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, W.C.; Zou, G.; Wang, G.S.; Kang, W.Y.; Yuan, J.; She, Z.G. Cytotoxic Bromine- and Iodine-Containing Cytochalasins Produced by the Mangrove Endophytic Fungus Phomopsis sp. QYM-13 Using the OSMAC Approach. J. Nat. Prod. 2022, 85, 1229–1238. [Google Scholar] [CrossRef]
- Zang, Z.M.; Yang, W.C.; Cui, H.; Cai, R.L.; Li, C.Y.; Zou, G.; Wang, B.; She, Z.G. Two Antimicrobial Heterodimeric Tetrahydroxanthones with a 7,7′-Linkage from Mangrove Endophytic Fungus Aspergillus flavus QQYZ. Molecules 2022, 27, 2691. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.M.; Cai, R.L.; Zang, Z.M.; Yang, W.C.; Wang, B.; Zhu, G.; Yuan, J.; She, Z.G. Azaphilone derivatives with anti-inflammatory activity from the mangrove endophytic fungus Penicillium sclerotiorum ZJHJJ-18. Bioorg. Chem. 2022, 122, 105721. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Liu, Z.M.; Chen, Y.C.; Tan, H.B.; Zhang, W.G.; Zhang, W.M. Polyketones from the endophytic fungus Cytospora rhizophorae. Nat. Prod. Res. 2021, 37, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Tan, H.B.; Wang, W.X.; Zhang, W.G.; Chen, Y.C.; Liu, S.N.; Liu, Z.M.; Li, H.H.; Zhang, W.M. Cytorhizophins A and B, benzophenone-hemiterpene adducts from the endophytic fungus Cytospora rhizophorae†. Org. Chem. Front. 2019, 6, 591–596. [Google Scholar] [CrossRef]
- Auamcharoen, W.; Kijjoa, A.; Chandrapatya, A.; Pinto, M.M.; Silva, A.M.S.; Naengchomnong, W.; Herz, W. A new tetralone from Diospyros cauliflora. Biochem. Syst. Ecol. 2009, 37, 690–692. [Google Scholar] [CrossRef]
- Machida, K.; Matsuoka, E.; Kasahara, T.; Kikuchi, M. Studies on the Constituents of Juglans Species. I. Structural Determination of (4S)- and (4R)-4-Hydroxy-α-tetralone Derivatives from the Fruit of Juglans mandshurica MAXIM. var. sieboldiana Makino. Chem. Pharm. Bull. 2005, 53, 934–937. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Yang, W.C.; Zou, G.; Chen, S.Y.; Pang, J.Y.; She, Z.G. Bioactive polyketides from the mangrove endophytic fungi Phoma sp. SYSUSK-7. Fitoterapia 2019, 139, 10436. [Google Scholar] [CrossRef]
- Qiu, P.; Liu, Z.M.; Chen, Y.; Cai, R.L.; Chen, G.Y.; She, Z.G. Secondary Metabolites with α-Glucosidase Inhibitory Activity from the Mangrove Fungus Mycosphaerella sp. SYSU-DZG01. Mar. Drugs 2019, 17, 483. [Google Scholar] [CrossRef] [Green Version]
- Ichikawa, K.; Sasada, R.; Chiba, K.; Gotoh, H. Effect of Side Chain Functional Groups on the DPPH Radical Scavenging Activity of Bisabolane-Type Phenols. Antioxidants 2019, 8, 65. [Google Scholar] [CrossRef] [Green Version]
- Odame, F.; Hosten, E.C.; Betz, R.; Krause, J.; Frost, C.L.; Lobb, K.; Tshentu, Z.R. Synthesis, characterization, computational studies and DPPH scavenging activity of some triazatetracyclic derivatives. J. Iran. Chem. Soc. 2021, 18, 1979–1995. [Google Scholar] [CrossRef]


| No. | 1 a | 2 b | 3 c | 4 c |
|---|---|---|---|---|
| 5 | 6.65, s | 6.54, s | 6.63, s | 6.58, s |
| 7 | 6.79, s | 6.82, s | 6.87, s | 6.75, s |
| 10 | 6.50, d (8.1) | 6.57, d (8.3) | 6.33, d (8.1) | 6.40, d (8.2) |
| 11 | 7.06, t, (8.1) | 7.15, t (8.3) | 6.99, t (8.1) | 7.02, t (8.2) |
| 12 | 6.50, d (8.1) | 6.57, d (8.3) | 6.33, d (8.1) | 6.40, d (8.2) |
| 14 | 2.24, s | 2.17, s | 2.27, s | 2.22, s |
| 1′ | 2.42, m | 2.42, t (7.1) | 3.41, s | |
| 2′ | 2.49, m | 2.66, t (7.1) | 7.30, m | |
| 3′ | 7.15, m | 7.09, m | ||
| 4′ | 3.58, s | 7.13, m | 7.07, m | |
| 5′ | 7.15, m | 7.02, t (8.2) | ||
| 6′ | 7.30, m | 7.07, m | ||
| 7′ | 7.09, m |
| No. | 1 a | 2 b | 3 c | 4 c |
|---|---|---|---|---|
| 1 | 198.2, C | 197.5, C | 199.1, C | 200.2, C |
| 2 | 134.4, C | 135.2, C | 133.6, C | 134.8, C |
| 3 | 152.1, C | 148.3, C | 154.0, C | 154.8, C |
| 3a | 127.1, C | 123.7, C | 127.1, C | 127.4, C |
| 4 | 151.7, C | 150.0, C | 153.1, C | 152.3, C |
| 5 | 124.5, CH | 124.7, CH | 125.1, CH | 125.0, CH |
| 6 | 140.7, C | 141.5, C | 141.9, C | 141.1, C |
| 7 | 116.4, CH | 118.2, CH | 116.9, CH | 116.6, CH |
| 7a | 134.3, C | 132.5, C | 134.4, C | 134.6, C |
| 8 | 110.6, C | 108.4, C | 112.1, C | 111.4, C |
| 9 | 156.2, C | 153.6, C | 156.5, C | 156.6, C |
| 10 | 108.0, CH | 109.4, CH | 107.9, CH | 107.8, CH |
| 11 | 130.7, CH | 131.9, CH | 130.7, CH | 130.8, CH |
| 12 | 108.0, CH | 109.4, CH | 107.9, CH | 107.8, CH |
| 13 | 156.2, C | 153.6, C | 156.5, C | 156.6, C |
| 14 | 21.0, CH3 | 21.3, CH3 | 21.2, CH3 | 21.1, CH3 |
| 1′ | 20.4, CH2 | 19.6, CH2 | 133.7, C | 30.5, CH2 |
| 2′ | 32.4, CH2 | 31.0, CH2 | 129.9, CH | 140.9, C |
| 3′ | 174.3, C | 175.1, C | 128.5, CH | 129.7, CH |
| 4′ | 52.2, CH3 | 127.9, CH | 128.9, CH | |
| 5′ | 128.5, CH | 126.5, CH | ||
| 6′ | 129.9, CH | 128.9, CH | ||
| 7′ | 129.7, CH |
| 6 a | ||
|---|---|---|
| No. | δC, Type | δH Mult (J in Hz) |
| 1 | 108.1, CH | 6.27, d (8.2) |
| 2 | 137.0, CH | 7.18, t (8.2) |
| 3 | 108.1, CH | 6.27, d (8.2) |
| 4 | 163.2, C | |
| 5 | 112.7, C | |
| 6 | 163.2, C | |
| 7 | 204.1, C | |
| 8 | 130.0, C | |
| 9 | 155.0, C | |
| 10 | 121.6, CH | 6.87, s |
| 11 | 140.8, C | |
| 12 | 121.6, CH | 7.25, s |
| 13 | 137.8, C | |
| 14 | 21.3, CH3 | 2.36, s |
| 15 | 202.2, C | |
| 16 | 38.9, CH2 | 2.96, t (7.5) |
| 17 | 20.6, CH2 | 1.85, m |
| 18 | 33.7, CH2 | 2.32, t (7.3) |
| 19 | 175.5, C | |
| 20 | 52.0, CH3 | 3.63, s |
| 7 a | ||
|---|---|---|
| No. | δC, Type | δH Mult (J in Hz) |
| 1 | 199.6, C | |
| 2 | 33.0, CH2 | 2.99, m |
| 2.43, dt (17.2, 3.6) | ||
| 3 | 31.6, CH2 | 2.26, m |
| 2.17, m | ||
| 4 | 61.7, CH | 5.26, t (3.1) |
| 4a | 139.9, C | |
| 5 | 146.2, C | |
| 6 | 157.5, C | |
| 7 | 117.8, C | 6.92, d (8.6) |
| 8 | 125.4, CH | 7.67, d (8.6) |
| 8a | 125.6, C | |
| 9 | 61.9, CH3 | |
| Compound | % Inhibition (100 µM) | EC50 (µM) |
|---|---|---|
| 1 | 90.8 | 11.5 ± 0.1 |
| 2 | 72.5 | 21.5 ± 1.0 |
| 3 | 69.0 | 19.7 ± 1.8 |
| 4 | 78.2 | 16.6 ± 0.4 |
| 5 | 81.0 | 12.4 ± 0.5 |
| 6 | 87.3 | 9.5 ± 0.1 |
| (+)-7 | 12.0 | – |
| (–)-7 | 4.2 | – |
| ascorbic acid a | 91.4 | 21.9 ± 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, G.; Li, T.; Yang, W.; Sun, B.; Chen, Y.; Wang, B.; Ou, Y.; Yu, H.; She, Z. Antioxidative Indenone and Benzophenone Derivatives from the Mangrove-Derived Fungus Cytospora heveae NSHSJ-2. Mar. Drugs 2023, 21, 181. https://doi.org/10.3390/md21030181
Zou G, Li T, Yang W, Sun B, Chen Y, Wang B, Ou Y, Yu H, She Z. Antioxidative Indenone and Benzophenone Derivatives from the Mangrove-Derived Fungus Cytospora heveae NSHSJ-2. Marine Drugs. 2023; 21(3):181. https://doi.org/10.3390/md21030181
Chicago/Turabian StyleZou, Ge, Taobo Li, Wencong Yang, Bing Sun, Yan Chen, Bo Wang, Yanghui Ou, Huijuan Yu, and Zhigang She. 2023. "Antioxidative Indenone and Benzophenone Derivatives from the Mangrove-Derived Fungus Cytospora heveae NSHSJ-2" Marine Drugs 21, no. 3: 181. https://doi.org/10.3390/md21030181
APA StyleZou, G., Li, T., Yang, W., Sun, B., Chen, Y., Wang, B., Ou, Y., Yu, H., & She, Z. (2023). Antioxidative Indenone and Benzophenone Derivatives from the Mangrove-Derived Fungus Cytospora heveae NSHSJ-2. Marine Drugs, 21(3), 181. https://doi.org/10.3390/md21030181