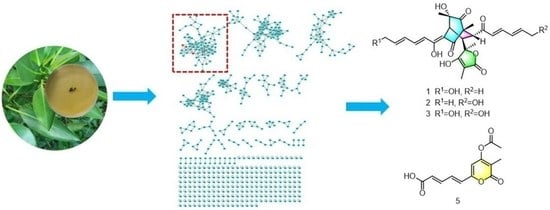
New Sorbicillinoids from the Mangrove Endophytic Fungus Trichoderma reesei SCNU-F0042
Abstract
:1. Introduction
2. Results
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. General Experimental Procedures
3.4. Spectral and Physical Data of Compounds 1–3 and 5
3.5. LC-MS/MS and Molecular Networking Analysis
3.6. SARS-CoV-2 Inhibition Assay
3.6.1. Cell Lines and Virus
3.6.2. Plasmids
3.6.3. RT-qPCR Analysis
3.7. Cell Viability Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Reino, J.L.; Guerrero, R.F.; Hernández-Galán, R.; Collado, I.G. Secondary Metabolites from Species of the Biocontrol Agent Trichoderma. Phytochem. Rev. 2008, 7, 89–123. [Google Scholar] [CrossRef]
- Chen, S.; Cai, R.; Liu, Z.; Cui, H.; She, Z. Secondary Metabolites from Mangrove-associated fungi: Source, Chemistry and Bioactivities. Nat. Prod. Rep. 2022, 39, 560–595. [Google Scholar] [CrossRef]
- Zhang, L.; Niaz, S.I.; Khan, D.; Wang, Z.; Zhu, Y.; Zhou, H.; Lin, Y.; Li, J.; Liu, L. Induction of Diverse Bioactive Secondary Metabolites from the Mangrove Endophytic Fungus Trichoderma sp. (Strain 307) by Co-Cultivation with Acinetobacter johnsonii (Strain B2). Mar. Drugs 2017, 15, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.-L.; Zhang, X.-F.; Huang, R.-H.; Wang, D.; Wang, X.-Q.; Li, Y.-Q.; Zheng, C.-J.; Zhang, P.; Zhang, C.-S. Antifungal Nafuredin and Epithiodiketopiperazine Derivatives from the Mangrove-Derived Fungus Trichoderma harzianum D13. Front. Microbiol. 2020, 11, 1495. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, J.-L.; Liu, J.-M.; Chen, R.-D.; Xie, K.-B.; Chen, D.-W.; Feng, K.-P.; Zhang, D.; Dai, J.-G. Neural Anti-Inflammatory Sesquiterpenoids from the Endophytic Fungus Trichoderma sp. Xy24. J. Asian Nat. Prod. Res. 2017, 19, 651–658. [Google Scholar] [CrossRef]
- Shiono, Y.; Miyazaki, N.; Murayama, T.; Koseki, T.; Harizon; Katja, D.G.; Supratman, U.; Nakata, J.; Kakihara, Y.; Saeki, M.; et al. GSK-3β Inhibitory Activities of Novel Dichroloresorcinol Derivatives from Cosmospora vilior Isolated from a Mangrove Plant. Phytochem. Lett. 2016, 18, 122–127. [Google Scholar] [CrossRef]
- Harned, A.M.; Volp, K.A. The Sorbicillinoid Family of Natural Products: Isolation, Biosynthesis, and Synthetic Studies. Nat. Prod. Rep. 2011, 28, 1790–1810. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yang, R.; Zheng, F.; Meng, X.; Zhang, W.; Liu, W. Dual Regulatory Role of Chromatin Remodeler ISW1 in Coordinating Cellulase and Secondary Metabolite Biosynthesis in Trichoderma reesei. Mbio 2022, 13, 03456. [Google Scholar] [CrossRef]
- Cram, D.J.; Tishler, M. Mold Metabolites; Isolation of Several Compounds from Clinical Penicillin. J. Am. Chem. Soc. 1948, 70, 4238. [Google Scholar] [CrossRef]
- Meng, J.; Wang, X.; Xu, D.; Fu, X.; Zhang, X.; Lai, D.; Zhou, L.; Zhang, G. Sorbicillinoids from Fungi and Their Bioactivities. Molecules 2016, 21, 715. [Google Scholar] [CrossRef] [Green Version]
- Rehman, S.; Yang, L.-J.; Zhang, Y.-H.; Wu, J.-S.; Shi, T.; Haider, W.; Shao, C.-L.; Wang, C.-Y. Sorbicillinoid Derivatives from Sponge-Derived Fungus Trichoderma reesei (HN-2016-018). Front. Microbiol. 2020, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wang, B.; Cheng, W. Study on the Secondary Metabolites of Thichoderma sturnisporum. Chin. J. Mar. Drugs 2017, 36, 27–31. [Google Scholar]
- Ngo, M.T.; Nguyen, M.V.; Han, J.W.; Park, M.S.; Kim, H.; Choi, G.J. In Vitro and In Vivo Antifungal Activity of Sorbicillinoids Produced by Trichoderma longibrachiatum. J. Fungi 2021, 7, 428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Deng, Y.; Lin, X.; Chen, B.; Li, J.; Liu, H.; Chen, S.; Liu, L. Anti-inflammatory Mono- and Dimeric Sorbicillinoids from the Marine-Derived Fungus Trichoderma reesei 4670. J. Nat. Prod. 2019, 82, 947–957. [Google Scholar] [CrossRef]
- Meng, J.; Gu, G.; Dang, P.; Zhang, X.; Wang, W.; Dai, J.; Liu, Y.; Lai, D.; Zhou, L. Sorbicillinoids from the Fungus Ustilaginoidea virens and Their Phytotoxic, Cytotoxic, and Antimicrobial Activities. Front. Chem. 2019, 7, 435. [Google Scholar] [CrossRef] [Green Version]
- Pang, X.; Zhou, X.; Lin, X.; Yang, B.; Tian, X.; Wang, J.; Xu, S.; Liu, Y. Structurally Various Sorbicillinoids from the Deep-Sea Sediment Derived Fungus Penicillium sp. SCSIO06871. Bioorg. Chem. 2021, 107, 104600. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; Felicio, R.; Fenner, A.; et al. Molecular Networking as a Dereplication Strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [Green Version]
- Andrade, R.; Ayer, W.A.; Trifonov, L.S. The Metabolites of Trichoderma longibrachiatum. III. Two New Tetronic Acids: 5-Hydroxyvertinolide and Bislongiquinolide. Aust. J. Chem. 1997, 50, 255–257. [Google Scholar] [CrossRef]
- Yu, J.; Han, H.; Zhang, X.; Ma, C.; Sun, C.; Che, Q.; Gu, Q.; Zhu, T.; Zhang, G.; Li, D. Discovery of Two New Sorbicillinoids by Overexpression of the Global Regulator LaeA in a Marine-Derived Fungus Penicillium dipodomyis YJ-11. Mar. Drugs 2019, 17, 446. [Google Scholar] [CrossRef] [Green Version]
- Han, W.; Cai, J.; Zhong, W.; Xu, G.; Wang, F.; Tian, X.; Zhou, X.; Liu, Q.; Liu, Y.; Wang, J. Protein Tyrosine Phosphatase 1B (PTP1B) Inhibitorsfrom the Deep-Sea Fungus Penicillium chrysogenum SCSIO 07007. Bioorg. Chem. 2020, 96, 103646. [Google Scholar] [CrossRef]
- Meng, J.; Cheng, W.; Heydari, H.; Wang, B.; Zhu, K.; Konuklugil, B.; Lin, W. Sorbicillinoid-Based Metabolites from a Sponge-Derived Fungus Trichoderma saturnisporum. Mar. Drugs 2018, 16, 226. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.-J.; Li, G.-H.; Shen, Y.-M. New Chemical Constituents from the Endophyte Streptomyces Species LR4612 Cultivated on Maytenus hookeri. Chem. Biodivers. 2006, 3, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.; Ayer, W.A.; Trifonov, L.S. The Metabolites of Trichoderma longibrachiatum. Part II The Structures of Trichodermolide and Sorbiquinol. Can. J. Chem. 1996, 74, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Abe, N.; Murata, T.; Hirota, A. Novel Oxidized Sorbicillin Dimers with 1,1-Diphenyl-2-Picrylhydrazyl-Radical Scavenging Activity from a Fungus. Biosci. Biotechnol. Biochem. 1998, 62, 2120–2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trifonov, L.S.; Hilpert, H.; Floersheim, P.; Dreiding, A.S.; Rast, D.M.; Skrivanova, R.; Hoesch, L. Bisvertinols: A New Group of Aimeric Vertinoids from Verticillium intertextum. Tetrahedron 1986, 42, 3157. [Google Scholar] [CrossRef]
- Kontani, M.; Sakagami, Y.; Marumo, S. First β-1,6-glucan biosynthesis inhibitor, bisvertinolone isolated from fungus, Acremonium strictum and its absolute stereochemistry. Tetrahedron Lett. 1994, 35, 2577. [Google Scholar] [CrossRef]
- Liu, J.; Gao, S.; Zhou, W.; Chen, Y.; Wang, Z.; Zeng, Z.; Zhou, H.; Lin, T. Dihydrotrichodimerol Purified from the Marine Fungus Acremonium citrinum Prevents NAFLD by Targeting PPARα. J. Nat. Prod. 2023, 86, 1189–1201. [Google Scholar] [CrossRef]
- Oguntuyo, K.Y.; Stevens, C.S.; Hung, C.; Ikegame, S.; Acklin, J.A.; Kowdle, S.S.; Carmichael, J.C.; Chiu, H.P.; Azarm, K.D.; Haas, G.D.; et al. Quantifying Absolute Neutralization Titers against SARS-CoV-2 by a Standardized Virus Neutralization Assay Allows for Cross-Cohort Comparisons of COVID-19 Sera. Mbio 2021, 12, 02492. [Google Scholar] [CrossRef]
- Tóth, G.; Horváti, K.; Kraszni, M.; Ausbüttel, T.; Pályi, B.; Kis, Z.; Mucsi, Z.; Kovács, G.M.; Bősze, S.; Boldizsár, I. Arylnaphthalene Lignans with Anti-SARS-CoV-2 and Antiproliferative Activities from the Underground Organs of Linum austriacum and Linum perenne. J. Nat. Prod. 2023, 86, 672–682. [Google Scholar] [CrossRef]





| NO | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH (J in Hz) | δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | δC, Type | |
| 1 | - | 63.7, C | - | 63.5, C | - | 63.6, C |
| 2 | - | 198.3, C | - | 197.5, C | - | 197.8, C |
| 3 | - | 111.7, C | - | 110.2, C | - | 110.5, C |
| 4 | 3.34, s | 43.6, CH | 3.33, s | 43.5, CH | 3.35, s | 43.6, CH |
| 5 | - | 75.9, C | - | 75.8, C | - | 75.8, C |
| 6 | - | 210.3, C | - | 210.2, C | - | 210.2, C |
| 7 | 3.36, d (5.6) | 52.2, CH | 3.31 a | 52.8, CH | 3.33 a | 52.6, CH |
| 8 | 3.22, d (5.6) | 43.9, CH | 3.28 a | 43.9, CH | 3.26 a | 43.9, CH |
| 9 | - | 168.6, C | - | 169.5, C | - | 169.4, C |
| 10 | 6.43–6.49 a | 121.6, CH | 6.28–6.33 | 119.6, CH | 6.43–6.46 | 121.5, CH |
| 11 | 7.35, dd (14.5, 11.2) | 142.5, CH | 7.30, dd (14.7, 11.2) | 143.7, CH | 7.37, dd (13.4, 11.5) | 142.7, CH |
| 12 | 6.55–6.59 a | 129.9, CH | 6.39–6.43 a | 132.4, CH | 6.52–6.60 a | 129.8, CH |
| 13 | 6.25–6.29 a | 143.0, CH | 6.19–6.25 a | 140.6, CH | 6.26–6.32 a | 143.3, CH |
| 14 | 4.22, d (4.4) | 63.0, CH2 | 1.90, d (6.4) | 18.9, CH3 | 4.23, d (4.1) | 62.8, CH2 |
| 15 | - | 203.1, C | - | 202.5, C | - | 202.1, C |
| 16 | 6.18, d (15.3) | 128.8, CH | 6.30–6.36 a | 130.0, CH | 6.31–6.40 a | 130.1, CH |
| 17 | 7.20, dd (15.3, 10.7) | 147.9, CH | 7.25, dd (15.4, 10.4) | 146.8, CH | 7.27, dd (15.2, 10.3) | 146.6, CH |
| 18 | 6.31–6.35 a | 131.7, CH | 6.48–6.53 a | 128.5, CH | 6.49–6.52 a | 128.6, CH |
| 19 | 6.38–6.42 a | 144.9, CH | 6.45–6.48 a | 147.6, CH | 6.48–6.51 a | 147.5, CH |
| 20 | 1.89, d (6.1) | 19.1, CH3 | 4.22, d (4.0) | 62.7, CH2 | 4.23, d (4.1) | 63.0, CH2 |
| 21 | - | 84.8, C | - | 84.6, C | - | 84.6, C |
| 22 | - | 185.5, C | - | 182.0, C | - | 182.9, C |
| 23 | - | 94.7, C | - | 96.2, C | - | 96.1, C |
| 24 | - | 179.1, C | - | 178.2, C | - | 178.2, C |
| 1-CH3 | 0.99, s | 11.4, CH3 | 0.99, s | 11.3, CH3 | 0.99, s | 11.3, CH3 |
| 5-CH3 | 1.18, s | 24.1, CH3 | 1.18, s | 24.2, CH3 | 1.19, s | 24.2, CH3 |
| 21-CH3 | 1.38, s | 23.3, CH3 | 1.40, s | 23.4, CH3 | 1.33, s | 23.3, CH3 |
| 23-CH3 | 1.45, s | 6.4, CH3 | 1.48, s | 6.5, CH3 | 1.47, s | 6.4, CH3 |
| NO | δH (J in Hz) | δC, Type |
|---|---|---|
| 2 | - | 163.6, C |
| 3 | - | 115.5, C |
| 4 | - | 158.9, C |
| 5 | 6.47, s | 106.6, CH |
| 6 | - | 155.6, C |
| 7 | - | 167.9, C |
| 8 | 2.34, s | 20.8, CH3 |
| 1′ | 6.70, d (15.2) | 130.2, CH |
| 2′ | 7.15, dd (15.2, 11.5) | 132.2, CH |
| 3′ | 7.37, dd (15.2, 11.5) | 143.4, CH |
| 4′ | 6.28, d (15.2) | 126.6, CH |
| 5′ | - | 167.4, C |
| 3-CH3 | 1.91, s | 10.3, CH3 |
| Compound | Inhibition of SARS-CoV-2 Viruses (EC50/μM) b | Cytotoxicity for 293T Cell (IC50/μM) b |
|---|---|---|
| 1 | NS c | >80 |
| 2 | 29.0 | >80 |
| 3 | NS | >80 |
| 5 | NS | >80 |
| Remdesivir a | 1.2 | >80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chen, T.; Yu, J.; Jia, H.; Chen, C.; Long, Y. New Sorbicillinoids from the Mangrove Endophytic Fungus Trichoderma reesei SCNU-F0042. Mar. Drugs 2023, 21, 442. https://doi.org/10.3390/md21080442
Li J, Chen T, Yu J, Jia H, Chen C, Long Y. New Sorbicillinoids from the Mangrove Endophytic Fungus Trichoderma reesei SCNU-F0042. Marine Drugs. 2023; 21(8):442. https://doi.org/10.3390/md21080442
Chicago/Turabian StyleLi, Jialin, Tao Chen, Jianchen Yu, Hao Jia, Chen Chen, and Yuhua Long. 2023. "New Sorbicillinoids from the Mangrove Endophytic Fungus Trichoderma reesei SCNU-F0042" Marine Drugs 21, no. 8: 442. https://doi.org/10.3390/md21080442
APA StyleLi, J., Chen, T., Yu, J., Jia, H., Chen, C., & Long, Y. (2023). New Sorbicillinoids from the Mangrove Endophytic Fungus Trichoderma reesei SCNU-F0042. Marine Drugs, 21(8), 442. https://doi.org/10.3390/md21080442