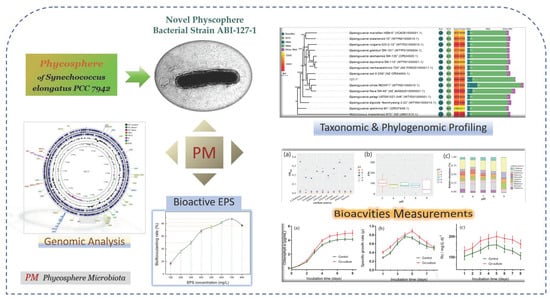
Taxonomic, Phylogenomic and Bioactivity Profiling of Novel Phycosphere Bacterium from Model Cyanobacterium Synechococcus elongatus PCC 7942
Abstract
:1. Introduction
2. Results and Discussion
2.1. Morphological and Biochemical Characterization of Strain ABI-127-1
2.2. Phylogenetic Characteristics of Strain ABI-127-1
2.3. Phylogenomic Inference
2.4. Genomic Sequencing and Annotation of Strain ABI-127-1
2.5. Biosynthetic Gene Prediction for Active Metabolites
2.6. Growth-Promoting Potential of Strain ABI-127-1 towards Algal Host
2.7. Culture Optimization for Bacterial EPS Production by Strain ABI-127-1
2.8. Bioflocculant Bioactivity of EPS Produced by Strain ABI-127-1
2.9. Bacterial Enchantment of Strain ABI-127-1 for Algal CO2 Fixation Efficiency
3. Materials and Methods
3.1. Bacterial Isolation and Culture Conditions
3.2. Phenotypic and Physiological Characterization
3.3. Phylogenetic Analysis
3.4. Genome Sequencing, Assembly, and Annotation
3.5. Phylogenomic Calculations
3.6. Multilocus Sequence Analysis (MLSA)
3.7. Bacterial Growth and EPS Production Measurements
3.8. Measurements of Bioflocculant and MGP Bioactivities
3.9. Measurement of Algal Carbon Fixation Efficiency in Co-Culture System
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giovannoni, S.J.; Nemergut, D. Microbes ride the current. Science 2014, 345, 1246. [Google Scholar] [CrossRef] [PubMed]
- Landry, Z.C.; Vergin, K.; Mannenbach, C.; Block, S.; Yang, Q.; Blainey, P.; Carlson, C.; Giovannoni, S.J. Optofluidic Single-Cell Genome Amplification of Sub-Micron Bacteria in the Ocean Subsurface. Front. Microbiol. 2018, 9, 1152. [Google Scholar] [CrossRef]
- Li, Z.; Li, S.; Chen, L.; Sun, T.; Zhang, W. Fast-growing cyanobacterial chassis for synthetic biology application. Crit. Rev. Biotechnol. 2023, 26, 1–15. [Google Scholar] [CrossRef]
- Doré, H.; Leconte, J.; Guyet, U.; Breton, S.; Farrant, G.K.; Demory, D.; Ratin, M.; Hoebeke, M.; Corre, E.; Pitt, F.D.; et al. Global Phylogeography of Marine Synechococcus in Coastal Areas Reveals Strong Community Shifts. mSystems 2022, 7, e0065622. [Google Scholar] [CrossRef]
- Díez, J.; López-Lozano, A.; Domínguez-Martín, M.A.; Gómez-Baena, G.; Muñoz-Marín, M.C.; Melero-Rubio, Y.; García-Fernández, J.M. Regulatory and metabolic adaptations in the nitrogen assimilation of marine picocyanobacteria. FEMS Microbiol. Rev. 2023, 47, fuac043. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed]
- Kouzuma, A.; Watanabe, K. Exploring the potential of algae/bacteria interactions. Curr. Opin. Biotechnol. 2015, 33, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Seymour, J.R.; Amin, S.A.; Raina, J.B.; Stocker, R. Zooming in on the phycosphere: The ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2017, 2, 17065. [Google Scholar] [CrossRef]
- Cirri, E.; Pohnert, G. Algae-bacteria interactions that balance the planktonic microbiome. New Phytol. 2019, 223, 100–106. [Google Scholar] [CrossRef]
- Yang, Q.; Ge, Y.M.; Iqbal, N.M.; Yang, X.; Zhang, X.L. Sulfitobacter alexandrii sp. nov.; a new microalgae growth-promoting bacterium with exopolysaccharides bioflocculanting potential isolated from marine phycosphere. Antonie Van Leeuwenhoek 2021, 114, 1091–1106. [Google Scholar] [CrossRef]
- Ramanan, R.; Kang, Z.; Kim, B.H.; Cho, D.H.; Jin, L.; Oh, H.M.; Kim, H.S. Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats. Algal Res. 2015, 8, 140–144. [Google Scholar] [CrossRef]
- Zhu, W.Z.; Ge, Y.M.; Dai, J.; Zhang, X.L.; Yang, Q. Alexandriicola marinus gen. nov.; sp. nov.; a new member of the family Rhodobacteraceae isolated from marine phycosphere. Antonie Van Leeuwenhoek 2022, 115, 473–486. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, D.; Nohynek, L.; Rischer, H. Algae-bacteria association inferred by 16S rDNA similarity in established microalgae cultures. MicrobiologyOpen 2014, 3, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Tian, X.Q.; Ma, L.Y.; Feng, B.; Liu, Q.H.; Yuan, L.D. Biodiversity of the symbiotic bacteria associated with toxic marine dinoflagellate Alexandrium tamarense. J. Biosci. Med. 2015, 3, 23–28. [Google Scholar] [CrossRef]
- Zhang, X.L.; Ma, L.Y.; Tian, X.Q.; Huang, H.L.; Yang, Q. Biodiversity study of intracellular bacteria closely associated with paralytic shellfish poisoning dinoflagellates Alexandrium tamarense and A. minutum. Int. J. Environ. Resour. 2015, 4, 23–27. [Google Scholar] [CrossRef]
- Feng, X.M.; Mo, Y.X.; Han, L.; Nogi, Y.; Zhu, Y.H.; Lv, J. Qipengyuania sediminis gen. nov.; sp. nov.; a member of the family Erythrobacteraceae isolated from subterrestrial sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 3658–3665. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.R.; Le, V.V.; Srivastava, A.; Kang, M.; Oh, H.M.; Ahn, C.Y. Algicidal activity of a novel bacterium, Qipengyuania sp. 3-20A1M, against harmful Margalefidinium polykrikoides: Effects of its active compound. Mar. Pollut. Bull. 2023, 186, 114397. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Garrity, G.M. Notification list. Notification that new names and new combinations have appeared in volume 70, part 8 of the IJSEM. Int. J. Syst. Evol. Microbiol. 2020, 70, 5602–5608. [Google Scholar]
- Zhao, Z.Y.; Xia, T.T.; Jiao, J.Y.; Liu, L.; Su, Q.Y.; Li, M.M.; Lv, A.P.; Ouyang, Y.T.; Li, W.J.; Ming, H. Qipengyuania thermophila sp. nov.; isolated from a Chinese hot spring. Arch. Microbiol. 2022, 204, 305. [Google Scholar] [CrossRef]
- Buchan, A.; González, J.M.; Moran, M.A. Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 2005, 71, 5665–5677. [Google Scholar] [CrossRef]
- Liu, Y.; Pei, T.; Du, J.; Yao, Q.; Deng, M.R.; Zhu, H. Comparative Genomics Reveals Genetic Diversity and Metabolic Potentials of the Genus Qipengyuania and Suggests Fifteen Novel Species. Microbiol. Spectr. 2022, 10, 0126421. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef]
- Li, S.; Liu, Q.; Duan, C.; Li, J.; Sun, H.; Xu, L.; Yang, Q.; Wang, Y.; Shen, X.; Zhang, L. c-di-GMP inhibits the DNA binding activity of H-NS in Salmonella. Nat. Commun. 2023, 14, 7502. [Google Scholar] [CrossRef] [PubMed]
- Dance, A. The search for microbial dark matter. Nature 2020, 582, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, S.; Liu, X.; Wang, Z.; Jiang, M.; Wang, R.; Xie, L.; Liu, Q.; Xie, X.; Shang, D.; et al. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 2020, 11, 5371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Li, G.X.; Ge, Y.M.; Iqbal, N.M.; Yang, X.; Cui, Z.D.; Yang, Q. Sphingopyxis microcysteis sp. nov.; a novel bioactive exopolysaccharides-bearing Sphingomonadaceae isolated from the Microcystis phycosphere. Antonie Van Leeuwenhoek 2021, 114, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.Z.; Gao, H.M.; Dai, J.; Zhu, W.Z.; Xu, F.F.; Ye, Y.; Zhang, X.L.; Yang, Q. Taxonomic and Bioactivity Characterizations of Mameliella alba Strain LZ-28 Isolated from Highly Toxic Marine Dinoflagellate Alexandrium catenella LZT09. Mar. Drugs 2022, 20, 321. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Feng, Q.; Zhang, B.P.; Gao, J.J.; Sheng, Z.; Xue, Q.P.; Zhang, X.L. Marinobacter alexandrii sp. nov.; a novel yellow-pigmented and algae growth-promoting bacterium isolated from marine phycosphere microbiota. Antonie Van Leeuwenhoek 2021, 114, 709–718. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, Z.; Zhou, X.; Zhang, R.; Xie, Z.; Zhang, S.; Wu, Y.; Ge, Y.; Zhang, X. Haliea alexandrii sp. nov.; isolated from phycosphere microbiota of the toxin-producing dinoflagellate Alexandrium catenella. Int. J. Syst. Evol. Microbiol. 2020, 70, 1133–1138. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, Z.; Zhou, X.; Zhang, R.; Wu, Y.; Lou, L.; Ma, Z.; Wang, D.; Ge, Y.; Zhang, X.; et al. Nioella ostreopsis sp. nov.; isolated from toxic dinoflagellate, Ostreopsis lenticularis. Int. J. Syst. Evol. Microbiol. 2020, 70, 759–765. [Google Scholar] [CrossRef]
- Sawa, N.; Tatsuke, T.; Ogawa, A.; Hirokawa, Y.; Osanai, T.; Hanai, T. Modification of carbon metabolism in Synechococcus elongatus PCC 7942 by cyanophage-derived sigma factors for bioproduction improvement. J. Biosci. Bioeng. 2019, 127, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, M.; Wang, M.; Zhang, W.; Qiao, C.; Luo, Q.; Lu, X. Freshwater Cyanobacterium Synechococcus elongatus PCC 7942 Adapts to an Environment with Salt Stress via Ion-Induced Enzymatic Balance of Compatible Solutes. Appl. Environ. Microbiol. 2020, 86, e02904-19. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, K.E.; Scaife, M.A.; Sasso, S.; Araujo, A.P.; Purton, S.; Smith, A.G. Unraveling vitamin B12-responsive gene regulation in algae. Plant Physiol. 2014, 165, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Croft, M.T.; Lawrence, A.D.; Raux-Deery, E.; Warren, M.J.; Smith, A.G. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 2005, 438, 90–93. [Google Scholar] [CrossRef]
- Guérin, H.; Kulakauskas, S.; Chapot-Chartier, M.P. Structural variations and roles of rhamnose-rich cell wall polysaccharides in Gram-positive bacteria. J. Biol. Chem. 2022, 298, 102488. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tomasch, J.; Jarek, M.; Wagner-Döbler, I. A dual-species co-cultivation system to study the interactions between Roseobacters and dinoflagellates. Front. Microbiol. 2014, 5, 311. [Google Scholar] [CrossRef]
- Islam, S.T.; Lam, J.S. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 2014, 60, 697–716. [Google Scholar] [CrossRef]
- Pérez-Burgos, M.; García-Romero, I.; Jung, J.; Schander, E.; Valvano, M.A.; Søgaard-Andersen, L. Characterization of the Exopolysaccharide Biosynthesis Pathway in Myxococcus xanthus. J. Bacteriol. 2020, 202, e00335-20. [Google Scholar] [CrossRef]
- Zhang, X.L.; Qi, M.; Li, Q.H.; Cui, Z.D.; Yang, Q. Maricaulis alexandrii sp. nov.; a novel active bioflocculants-bearing and dimorphic prosthecate bacterium isolated from marine phycosphere. Antonie Van Leeuwenhoek 2021, 114, 1195–1203. [Google Scholar] [CrossRef]
- Bruckner, C.G.; Rehm, C.; Grossart, H.P.; Kroth, P.G. Growth and release of extracellular organic compounds by benthic diatoms depend on interactions with bacteria. Environ. Microbiol. 2011, 13, 1052–1063. [Google Scholar] [CrossRef]
- Daly, G.; Perrin, E.; Viti, C.; Fondi, M.; Adessi, A. Scaling down the microbial loop: Data-driven modelling of growth interactions in a diatom-bacterium co-culture. Environ. Microbiol. Rep. 2021, 13, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Mistou, M.Y.; Sutcliffe, I.C.; van Sorge, N.M. Bacterial glycobiology: Rhamnose- containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol. Rev. 2016, 40, 464–479. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Chen, B.; Yang, Y.; Yi, X.; Wei, M.; Ecklu-Mensah, G.; Buschmann, M.M.; Liu, H.; Gao, J.; Liang, W.; et al. Multi-omics analyses of airway host-microbe interactions in chronic obstructive pulmonary disease identify potential therapeutic interventions. Nat. Microbiol. 2022, 7, 1361–1375. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, J.; Ge, Y.; Yang, Q.; Sun, J.; Yu, X. Acute effects of CH3NH3PbI3 perovskite on Scenedesmus obliquus and Daphnia magana in aquatic environment. Ecotoxicol. Environ. Saf. 2021, 208, 111677. [Google Scholar] [CrossRef] [PubMed]
- Barott, K.L.; Rohwer, F.L. Unseen players shape benthic competition on coral reefs. Trends Microbiol. 2012, 20, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, Z.; Zhou, X.; Xie, Z.; Wang, Y.; Wang, D.; Feng, L.; Yang, G.; Ge, Y.; Zhang, X. Saccharospirillum alexandrii sp. nov.; isolated from the toxigenic marine dinoflagellate Alexandrium catenella LZT09. Int. J. Syst. Evol. Microbiol. 2020, 70, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Jiang, Z.; Wu, Z.; Sheng, Z.; Yang, X.; Sun, J.; Zhang, X.; Yang, Q.; Yu, X.; Yan, J. Limnobacter alexandrii sp. nov.; a thiosulfate-oxidizing, heterotrophic and EPS-bearing Burkholderiaceae isolated from cultivable phycosphere microbiota of toxic Alexandrium catenella LZT09. Antonie Van Leeuwenhoek 2020, 13, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Yang, X.; Wang, S.J.; Jiang, Z.W.; Xie, Z.X.; Zhang, L. Draft Genome Sequences of Nine Cultivable Heterotrophic Proteobacteria Isolated from Phycosphere Microbiota of Toxic Alexandrium catenella LZT09. Microbiol. Resour. Announc. 2020, 9, e00281-20. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, X.; Jiang, Z.; Yang, X.; Zhang, X.; Yang, Q. Combined characterization of a new member of Marivita cryptomonadis, strain LZ-15-2 isolated from cultivable phycosphere microbiota of toxic HAB dinoflagellate Alexandrium catenella LZT09. Braz. J. Microbiol. 2021, 52, 739–748. [Google Scholar] [CrossRef]
- Gao, H.M.; Xie, P.F.; Zhang, X.L.; Yang, Q. Isolation, Phylogenetic and Gephyromycin Metabolites Characterization of New Exopolysaccharides-Bearing Antarctic Actinobacterium from Feces of Emperor Penguin. Mar. Drugs 2021, 19, 458. [Google Scholar] [CrossRef]
- Yang, X.; Jiang, Z.W.; Zhang, J.; Zhou, X.; Zhang, X.L.; Wang, L.; Yu, T.; Wang, Z.; Bei, J.; Dong, B. Mesorhizobium alexandrii sp. nov.; isolated from phycosphere microbiota of PSTs-producing marine dinoflagellate Alexandrium minutum amtk4. Antonie Van Leeuwenhoek 2020, 113, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Duan, Y.; Yang, X.; Yao, B.; Zeng, T.; Wang, X.; Feng, Q.; Qi, M.; Yang, Q.; Zhang, X.L. Nitratireductor alexandrii sp. nov.; from phycosphere microbiota of toxic marine dinoflagellate Alexandrium tamarense. Int. J. Syst. Evol. Microbiol. 2020, 70, 4390–4397. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, Z.W.; Huang, C.H.; Zhang, R.N.; Li, L.Z.; Yang, G.; Feng, L.J.; Yang, G.F.; Zhang, H.; Zhang, X.L. Hoeflea prorocentri sp. nov.; isolated from a culture of the marine dinoflagellate Prorocentrum mexicanum PM01. Antonie Van Leeuwenhoek 2018, 111, 1845–1853. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Philippot, L.; Andersson, S.G.; Battin, T.J.; Prosser, J.I.; Schimel, J.P.; Whitman, W.B.; Hallin, S. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 2010, 8, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, S.P.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.; Lee, Y.C. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef]
- Siddharth, T.; Sridhar, P.; Vinila, V.; Tyagi, R.D. Environmental applications of microbial extracellular polymeric substance (EPS): A review. J. Environ. Manag. 2021, 287, 112307. [Google Scholar] [CrossRef]
- Gonzalez, L.E.; Bashan, Y. Increased growth of the microalga Chlorella vulgaris when coimmobilized and cocultured in alginate beads with the plant-growth-promoting bacterium Azospirillum brasilense. Appl. Environ. Microbiol. 2000, 6, 1527–1531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, W.; Li, Q.; Cui, R.; Wang, Z.; Wang, Y.; Zhang, Y.Z.; Ding, W.; Shen, X. Deciphering the Root Endosphere Microbiome of the Desert Plant Alhagi sparsifolia for Drought Resistance-Promoting Bacteria. Appl. Environ. Microbiol. 2020, 86, e02863-19. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, L.; Zhang, H.; Liu, Z.; An, F.; Yang, Q.; Zhang, X.; Yao, L.; Yang, S.; Dai, J.; Chen, X. Improved Glutamic Acid Production Capacity of Corynebacterium glutamicum by the ARTP Mutagenesis Method. Fermentation 2023, 9, 599. [Google Scholar] [CrossRef]









| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Isolation source | marine phycosphere | seawater | seawater | seawater | seawater | marine sediment |
| Colony color | Yellow | Yellow | Orange | Orange | Orange | Orange |
| Motility | − | − | − | − | − | + |
| Nitrate reduction | + | + | − | − | + | − |
| Tolerance to NaCl (%, v/v, optimal) | 1.0–10.0 (2.5) | 1.0–10.0 (3.0) | 1.0–9.0 (2–3) | 0.5–15.0 (3.0) | 0.5–9.0 (2–3) | 0–10.0 (3.5) |
| Growth temperature (°C, optimal) | 15–35 (28–30) | 4–37 (25–30) | 10–40 (30–35) | 10–41 (30–35) | 12–40 (30–36) | 10-40 (30–35) |
| Growth pH range (optimal) | 5–10 (7–8) | 5–10 (7–8) | 5–10 (7–8) | 5–10 (7–8) | 5–10 (8–9) | 5–10 (7–8) |
| Utilization of | ||||||
| Glucose | − | + | + | + | + | + |
| Fructose | + | − | − | − | − | − |
| Lactate | + | − | − | − | − | − |
| 16S rRNA gene similarity (%) | − | 98.64 | 97.79 | 98.29 | 97.29 | 97.79 |
| DNA G+C content (mol%) | 62.15 | 62.01 | 62.18 | 62.17 | 60.40 | 59.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Xu, J.; Dai, J.; Zhang, L.; Feng, L.; Tian, X.; Yang, Q. Taxonomic, Phylogenomic and Bioactivity Profiling of Novel Phycosphere Bacterium from Model Cyanobacterium Synechococcus elongatus PCC 7942. Mar. Drugs 2024, 22, 36. https://doi.org/10.3390/md22010036
Zhang X, Xu J, Dai J, Zhang L, Feng L, Tian X, Yang Q. Taxonomic, Phylogenomic and Bioactivity Profiling of Novel Phycosphere Bacterium from Model Cyanobacterium Synechococcus elongatus PCC 7942. Marine Drugs. 2024; 22(1):36. https://doi.org/10.3390/md22010036
Chicago/Turabian StyleZhang, Xiaoling, Jiaquan Xu, Jun Dai, Lei Zhang, Lijuan Feng, Xiaoqing Tian, and Qiao Yang. 2024. "Taxonomic, Phylogenomic and Bioactivity Profiling of Novel Phycosphere Bacterium from Model Cyanobacterium Synechococcus elongatus PCC 7942" Marine Drugs 22, no. 1: 36. https://doi.org/10.3390/md22010036
APA StyleZhang, X., Xu, J., Dai, J., Zhang, L., Feng, L., Tian, X., & Yang, Q. (2024). Taxonomic, Phylogenomic and Bioactivity Profiling of Novel Phycosphere Bacterium from Model Cyanobacterium Synechococcus elongatus PCC 7942. Marine Drugs, 22(1), 36. https://doi.org/10.3390/md22010036