Cloning and Characterization of a Novel N-Acetyl-D-galactosamine-4-O-sulfate Sulfatase, SulA1, from a Marine Arthrobacter Strain
Abstract
:1. Introduction
2. Results
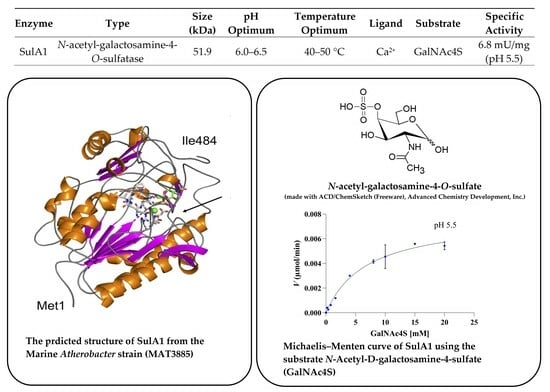
2.1. Phylogenetic Identification, Identification of SulA1, and Modelling of the Enzyme
2.2. Recombinant Production in E. coli, and Purification of SulA1
2.3. The pH and Temperature Profile of SulA1
2.4. Storage Stability
2.5. Cation Dependence
2.6. Sulfatase Activity against Natural Substrates
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Plasmids
4.2. Sequencing, Bioinformatic Analysis, and Structure Modeling
Substrate Docking and MD Simulation
4.3. Cloning of the Sulfatase Gene sulA1
4.4. Production, Purification, and Storage of the Recombinant Sulfatase
4.5. Total Protein Content and Protein Distribution
4.6. Sulfatase Activity—Standard Assay
4.7. Temperature Optimum, Thermal Deactivation, and Storage Capacity
4.8. pH Optimum
4.9. The Effect of Cations and EDTA on Activity
4.10. Differential Scanning Fluorimetry (DSF) in Presence and Absence of Calcium Ions and EDTA
4.11. Peptide Mass Fingerprinting
4.12. Preparation of Sulfated Mono-, Di-, and Oligosaccharide Solutions from Various Resources
4.13. Analysis of the Degradation Product by Thin-Layer Chromatography
4.14. Determination of the Kinetic Properties and the Specific Activity of SulA1
4.15. Graphs and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silchenko, A.S.; Rasin, A.B.; Zueva, A.O.; Kusaykin, M.I.; Zvyagintseva, T.N.; Kalinovsky, A.I.; Kurilenko, V.V.; Ermakova, S.P. Fucoidan Sulfatases from Marine Bacterium Wenyingzhuangia Fucanilytica CZ1127T. Biomolecules 2018, 8, 98. [Google Scholar] [CrossRef]
- Wang, S.; Su, T.; Zhang, Q.; Guan, J.; He, J.; Gu, L.; Li, F. Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases with High Identity. Front. Microbiol. 2019, 10, 1309. [Google Scholar] [CrossRef]
- Stam, M.; Lelièvre, P.; Hoebeke, M.; Corre, E.; Barbeyron, T.; Michel, G. SulfAtlas, the Sulfatase Database: State of the Art and New Developments. Nucleic Acids Res. 2023, 51, D647–D653. [Google Scholar] [CrossRef] [PubMed]
- Barbeyron, T.; Brillet-Guéguen, L.; Carré, W.; Carrière, C.; Caron, C.; Czjzek, M.; Hoebeke, M.; Michel, G. Matching the Diversity of Sulfated Biomolecules: Creation of a Classification Database for Sulfatases Reflecting Their Substrate Specificity. PLoS ONE 2016, 11, e0164846. [Google Scholar] [CrossRef]
- Helbert, W. Marine Polysaccharide Sulfatases. Front. Mar. Sci. 2017, 4, 6. [Google Scholar] [CrossRef]
- Wang, S.; Sugahara, K.; Li, F. Chondroitin Sulfate/Dermatan Sulfate Sulfatases from Mammals and Bacteria. Glycoconj. J. 2016, 33, 841–851. [Google Scholar] [CrossRef]
- Wei, L.; Zhang, Q.; Lu, D.; Du, M.; Xu, X.; Wang, W.; Zhang, Y.Z.; Yuan, X.; Li, F. Identification and Action Patterns of Two Chondroitin Sulfate Sulfatases From a Marine Bacterium Photobacterium Sp. QA16. Front. Microbiol. 2022, 12, 775124. [Google Scholar] [CrossRef] [PubMed]
- Hanson, S.R.; Best, M.D.; Wong, C.H. Sulfatases: Structure, Mechanism, Biological Activity, Inhibition, and Synthetic Utility. Angew. Chem. Int. Ed. 2004, 43, 5736–5763. [Google Scholar] [CrossRef]
- Schlachter, C.R.; Malley, A.O.; Grimes, L.L.; Tomashek, J.J.; Chruszcz, M.; Lee, L.A. Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter Yulinensis. Molecules 2022, 27, 87. [Google Scholar] [CrossRef]
- Wang, S.; Guan, J.; Zhang, Q.; Chen, X.; Li, F. Identification and Signature Sequences of Bacterial Delta(4,5)Hexuronate-2-O-Sulfatases. Front. Microbiol. 2019, 10, 704. [Google Scholar] [CrossRef]
- Myette, J.R.; Shriver, Z.; Claycamp, C.; Mclean, M.W.; Venkataraman, G.; Sasisekharan, R. The Heparin/Heparan Sulfate 2-O-Sulfatase from Flavobacterium Heparinum. J. Biol. Chem. 2003, 278, 12157–12166. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, J.E.; Vilén, E.M.; Namburi, R.B.; Benjdia, A.; Beneteau, J.; Malleron, A.; Bonnaffé, D.; Driguez, P.A.; Descroix, K.; Lassalle, G.; et al. Characterization of Glycosaminoglycan (GAG) Sulfatases from the Human Gut Symbiont Bacteroides Thetaiotaomicron Reveals the First GAG-Specific Bacterial Endosulfatase. J. Biol. Chem. 2014, 289, 24289–24303. [Google Scholar] [CrossRef]
- Wang, W.; Han, W.; Cai, X.; Zheng, X.; Sugahara, K.; Li, F. Cloning and Characterization of a Novel Chondroitin Sulfate/Dermatan Sulfate 4-O-Endosulfatase from a Marine Bacterium. J. Biol. Chem. 2015, 290, 7823–7832. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, T.; Saito, H.; Habuchi, O.; Suzuki, S. Purification and Properties of Bacterial Chondroitinases and Chondrosulfatases. J. Biol. Chem. 1968, 243, 1523–1535. [Google Scholar] [CrossRef]
- Mucci, A.; Schenetti, L.; Volpi, N. 1H and 13 C Nuclear Magnetic Resonance Identification and Characterization of Components of Chondroitin Sulfates of Various Origin. Carbohydr. Polym. 2000, 41, 37–45. [Google Scholar] [CrossRef]
- Abdallah, M.M.; Fernández, N.; Matias, A.A.; Bronze, M. do R. Hyaluronic Acid and Chondroitin Sulfate from Marine and Terrestrial Sources: Extraction and Purification Methods. Carbohydr. Polym. 2020, 243, 116441. [Google Scholar] [CrossRef]
- Chen, S.; Xue, C.; Yin, L.; Tang, Q.; Yu, G.; Chai, W. Comparison of Structures and Anticoagulant Activities of Fucosylated Chondroitin Sulfates from Different Sea Cucumbers. Carbohydr. Polym. 2011, 83, 688–696. [Google Scholar] [CrossRef]
- García, A.G.; Vergés, J.; Montell, E. Immunomodulatory and Anti-Inflammatory Effects of Chondroitin Sulphate. J. Cell Mol. Med. 2009, 13, 1451–1463. [Google Scholar] [CrossRef]
- Ustyuzhanina, N.E.; Bilan, M.I.; Dmitrenok, A.S.; Shashkov, A.S. Two Structurally Similar Fucosylated Chondroitin Sulfates from the Holothurian Species Stichopus Chloronotus and Stichopus Horrens. Carbohydr. Polym. 2018, 189, 10–14. [Google Scholar] [CrossRef]
- Ilieva, K.M.; Cheung, A.; Mele, S.; Chiaruttini, G.; Crescioli, S.; Griffin, M.; Nakamura, M.; Spicer, J.F.; Tsoka, S.; Lacy, K.E.; et al. Chondroitin Sulfate Proteoglycan 4 and Its Potential As an Antibody Immunotherapy Target across Different Tumor Types. Front. Immunol. 2018, 8, 1911. [Google Scholar] [CrossRef]
- Bougatef, H.; Krichen, F.; Capitani, F.; Amor, I.B.; Maccari, F.; Mantovani, V.; Galeotti, F.; Volpi, N.; Bougatef, A.; Sila, A. Chondroitin Sulfate/Dermatan Sulfate from Corb (Sciaena Umbra) Skin: Purification, Structural Analysis and Anticoagulant Effect. Carbohydr. Polym. 2018, 196, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Randomized, A.; Gabay, C.; Medinger-sadowski, C.; Gascon, D.; Kolo, F.; Finckh, A. Symptomatic Effects of Chondroitin 4 and Chondroitin 6 Sulfate on Hand Osteoarthritis. Arthritis Rheum. 2011, 63, 3383–3391. [Google Scholar] [CrossRef]
- Kale, V.; Friðjónsson, Ó.; Jónsson, J.Ó.; Kristinsson, H.G.; Ómarsdóttir, S.; Hreggviðsson, G. Chondroitin Lyase from a Marine Arthrobacter Sp. MAT3885 for the Production of Chondroitin Sulfate Disaccharides. Mar. Biotechnol. 2015, 17, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Zou, X. Advances and Challenges in Protein-Ligand Docking. Int. J. Mol. Sci. 2010, 11, 3016–3034. [Google Scholar] [CrossRef] [PubMed]
- Appel, M.J.; Bertozzi, C.R. Formylglycine, a Post-Translationally Generated Residue with Unique Catalytic Capabilities and Biotechnology Applications. ACS Chem. Biol. 2015, 10, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Ndeh, D.; Baslé, A.; Strahl, H.; Yates, E.A.; McClurgg, U.L.; Henrissat, B.; Terrapon, N.; Cartmell, A. Metabolism of Multiple Glycosaminoglycans by Bacteroides Thetaiotaomicron Is Orchestrated by a Versatile Core Genetic Locus. Nat. Commun. 2020, 11, 646. [Google Scholar] [CrossRef]
- Matusiewicz, M.; Krzystek-Korpacka, M.; Dabrowski, K. Characterization of Arylsulfatase Activity in Brine Shrimp, Artemia Salina. J. Exp. Mar. Biol. Ecol. 2005, 317, 175–187. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Look, D.; Tobacman, J.K. Increased Arylsulfatase B Activity in Cystic Fibrosis Cells Following Correction of CFTR. Clin. Chim. Acta 2007, 380, 122–127. [Google Scholar] [CrossRef]
- Berteau, O.; Guillot, A.; Benjdia, A.; Rabot, S. A New Type of Bacterial Sulfatase Reveals a Novel Maturation Pathway in Prokaryotes. J. Biol. Chem. 2006, 281, 22464–22470. [Google Scholar] [CrossRef] [PubMed]
- Sardiello, M.; Annunziata, I.; Roma, G.; Ballabio, A. Sulfatases and Sulfatase Modifying Factors: An Exclusive and Promiscuous Relationship. J. Jpn. For. Soc. 2005, 14, 3203–3217. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.D.; William, D.L.; Janet, G.H. Comparative Studies of Rodent Anionic Arylsulfatases. Comp. Biochem. Physiol. Part B Biochem. 1985, 82, 55–61. [Google Scholar] [CrossRef]
- Wegerer, A.; Sun, T.; Altenbuchner, J. Optimization of an E. coli L-Rhamnose-Inducible Expression Vector: Test of Various Genetic Module Combinations. BMC Biotechnol. 2008, 8, 2. [Google Scholar] [CrossRef]
- Dumon-Seignovert, L.; Cariot, G.; Vuillard, L. The Toxicity of Recombinant Proteins in Escherichia coli: A Comparison of Overexpression in BL21(DE3), C41(DE3), and C43(DE3). Protein Expr. Purif. 2004, 37, 203–206. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Land, H.; Humble, M.S. YASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations. Methods Mol. Biol. 2018, 1685, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Steinegger, M.; Söding, J. MMseqs2 Desktop and Local Web Server App for Fast, Interactive Sequence Searches. Bioinformatics 2019, 35, 2856–2858. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Dower, W.J.; Miller, J.F.; Ragsdale, C.W. High Efficiency Transformation of E. coli by High Voltage Electroporation. Nucleic Acids Res. 1988, 16, 6127–6145. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophor. Int. J. 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havliš, J.; Olsen, J.V.; Mann, M. In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 2007, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Jork, H.; Funk, W.; Fischer, W.; Wimmer, H. Thin-Layer Chromatography; VCH: Weinheim, Germany, 1990; Volume 1a, ISBN 3527278346. [Google Scholar]









| Template (PDB Code) | Source | Sequence Coverage (%) | Amino Acid Sequence Identity (%) | Transferred Region to the Model (Residue Numbering) |
|---|---|---|---|---|
| 5G2V | Bacteroides thetaiotaomicron VPI-5482 | 76 | 32.09 | 1–459 |
| 6B0K | Pseudoalteromonas sp. | 89 | 27.27 | 168–180, 435–483 |
| 2QZU | Bacteroides fragilis YCH46 | 90 | 24.79 | 1–9, 283–282, 433–441 |
| 6BIA | Pseudoalteromonas fuliginea | 90 | 27.20 | 452–484 |
| 6UST | Hungatella hathewayi | 94 | 30.60 | 86–89 |
| Check Type | Quality Z-Score | Comment |
|---|---|---|
| Dihedrals | −0.660 | Good |
| Packing 1D | −1.453 | Satisfactory |
| Packing 3D | −2.417 | Poor |
| Overall | −1.786 | Satisfactory |
| Sample | Relative Activity (%) |
|---|---|
| Crude SulA1 | 24.4 ± 1.08 |
| Pure SulA1 | 100.0 ± 0.0 |
| CaCl2 (mM) | Tm (°C) | EDTA (mM) | Tm (°C) |
|---|---|---|---|
| 0 | 67.2 ± 0.15 | 0 | 67.5 ± 0.09 |
| 7 | 64.0 ± 0.06 | 0.5 | 67.3 ± 0.21 |
| 14 | 62.3 ± 0.10 | 1 | 67.3 ± 0.67 |
| 2 | 67.1 ± 0.54 | ||
| 5 | 67.0 ± 0.15 |
| Substrates | Activity |
|---|---|
| Chondroitin sulfate AC dp4 (contain both 4S and 6S) | − |
| Chondroitin without sulfation (Δdi0S) | − |
| Chondroitin sulfate (Δdi4S) | − |
| Chondroitin sulfate (Δdi6S) | − |
| N-Acetyl-D-galactosamine-4-sulfate (GalNAc4S) | + |
| N-Acetyl-D-galactosamine-6-sulfate (GalNAc6S) | − |
| Heparin disaccharide I-H | − |
| N-Acetyl-D-glucosamine-6-sulfate (GlcNac-6S) | − |
| D-galactose-4-sulfate (Gal-4S) | − |
| Neocarrabiose-4-O-sulfate | − |
| Neocarratetraose-41,43-di-O-sulfate | − |
| L-fucose from Chorda filum cut-off <10 kDa Fuc310) | − |
| Fucoidan | − |
| Enzyme | Type | Size (kDa) | pH Optimum | Temperature Optimum | Ligand | Substrate | Specific Activity | Reference |
|---|---|---|---|---|---|---|---|---|
| SulA1 | N-acetyl-galactosamine-4-O-sulfatase | 51.9 | 6.0–6.5 | 40–50 °C | Ca2+ | GalNAc4S | pH 5.5 6.8 mU/mg (at 20 mM GalNAc4S) pH 6.5 14.0 mU/mg (at 15 mM GalNAc4S) | This work |
| endoVB4SF (Vibrio sp. FC509)–pdb:6J66_A | CS/DS 4-O-endosulfatase | 59.5 | 8.0 | 30 °C | none | ΔA 1 CS-A | 5020 and 820 mU/mg | [13] |
| exoPB4SF (Photobacterium sp.)–QAB47431.1 | Exolytic N-acetyl-galactosamine-4-O-sulfatase | 56.1 | 8.0 | 30 °C | Na+, Li+, K+, Mg2+, and Ca2+ | ΔA | 2857 mU/mg (for the purified protein) | [2] |
| PB_3262 (Photobacterium sp. QA16)–UF91287.1 | Endo-CS-4-O-Sulfatase | 56.7 | 7.0 | 50 °C | Ca2+ Ba2+ | ΔA | 165.750 U/mg | [7] |
| PB_3285 (Photobacterium sp. QA16)–UF91288.1 | Exolytic CS N-acetyl-galactosamine-6-O-sulfatase | 57.3 | 7.0 | 30 °C | Ca2+ K+ | ΔC 2 | 3.64 mU/mg | [7] |
| Parameter | Description |
|---|---|
| Modeling speed | Slow |
| PSI-BLAST iterations in template search | 3 |
| Maximum allowed (PSI-)BLAST E-value to consider template (EValue Max) | 0.5 |
| Maximum number of templates to be used | 5 |
| Maximum number of templates with same sequence | 1 |
| Maximum oligomerization state | 4 |
| Maximum number of alignment variations per template | 5 |
| Maximum number of conformations tried per loop | 50 |
| Maximum number of residues added to the termini | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christensen, M.D.; Allahgholi, L.; Linares-Pastén, J.A.; Friðjónsson, Ó.; Guðmundsson, H.; Kale, V.; Sardari, R.R.R.; Hreggviðsson, G.Ó.; Karlsson, E.N. Cloning and Characterization of a Novel N-Acetyl-D-galactosamine-4-O-sulfate Sulfatase, SulA1, from a Marine Arthrobacter Strain. Mar. Drugs 2024, 22, 104. https://doi.org/10.3390/md22030104
Christensen MD, Allahgholi L, Linares-Pastén JA, Friðjónsson Ó, Guðmundsson H, Kale V, Sardari RRR, Hreggviðsson GÓ, Karlsson EN. Cloning and Characterization of a Novel N-Acetyl-D-galactosamine-4-O-sulfate Sulfatase, SulA1, from a Marine Arthrobacter Strain. Marine Drugs. 2024; 22(3):104. https://doi.org/10.3390/md22030104
Chicago/Turabian StyleChristensen, Monica Daugbjerg, Leila Allahgholi, Javier A. Linares-Pastén, Ólafur Friðjónsson, Hörður Guðmundsson, Varsha Kale, Roya R. R. Sardari, Guðmundur Ó. Hreggviðsson, and Eva Nordberg Karlsson. 2024. "Cloning and Characterization of a Novel N-Acetyl-D-galactosamine-4-O-sulfate Sulfatase, SulA1, from a Marine Arthrobacter Strain" Marine Drugs 22, no. 3: 104. https://doi.org/10.3390/md22030104
APA StyleChristensen, M. D., Allahgholi, L., Linares-Pastén, J. A., Friðjónsson, Ó., Guðmundsson, H., Kale, V., Sardari, R. R. R., Hreggviðsson, G. Ó., & Karlsson, E. N. (2024). Cloning and Characterization of a Novel N-Acetyl-D-galactosamine-4-O-sulfate Sulfatase, SulA1, from a Marine Arthrobacter Strain. Marine Drugs, 22(3), 104. https://doi.org/10.3390/md22030104