A Tropical Marine Microbial Natural Products Geobibliography as an Example of Desktop Exploration of Current Research Using Web Visualisation Tools
Abstract
: Microbial marine biodiscovery is a recent scientific endeavour developing at a time when information and other technologies are also undergoing great technical strides. Global visualisation of datasets is now becoming available to the world through powerful and readily available software such as Worldwind™, ArcGIS Explorer™ and Google Earth™. Overlaying custom information upon these tools is within the hands of every scientist and more and more scientific organisations are making data available that can also be integrated into these global visualisation tools. The integrated global view that these tools enable provides a powerful desktop exploration tool. Here we demonstrate the value of this approach to marine microbial biodiscovery by developing a geobibliography that incorporates citations on tropical and near-tropical marine microbial natural products research with Google Earth™ and additional ancillary global data sets. The tools and software used are all readily available and the reader is able to use and install the material described in this article.- If not already done, install Google Earth™ free version from http://earth.google.com/
- Download file “Tropical marine microbial natural products geo bibliography.kml”
- Download file “Supporting KML files marine microbial natural products geobibliography.kmz”
- Start Google Earth™
- Use File>Open in the menu and open the abovementioned files
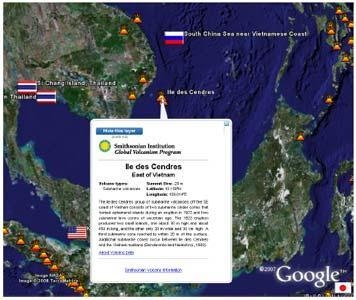
- A dataset list appears in a window on the right hand side of the screen enabling the reader to switch on or off the display of individual datasets
- Some of the datasets require active internet connections to download the required information and the required data may take significant time to download during which time a small moving icon is often displayed by Google Earth™
1. Introduction
2. Materials and Methods
Google Earth™
Making the geobibliography
- - the original microbial collection site was reported or could be determined from the article.
- - the collection site was tropical or near-tropical
- - the means of producing the natural product was primarily by culture of the micro-organisms rather than by simply collecting large wild populations (eg cyanobacterial mats or microscopic blooms)
- - a name for the location;
- - the citation for the work;
- - a link to the publication’s digital object identifier (DOI), or in those few cases where a DOI was unavailable, directly to the article if it were freely available;
- - a link to a PubChem [91] entry if it existed;
- - longitude and latitude converted to decimal format. For those readers who examine the KML file with a text editor, note that the third number that appears after the longitude and latitude defines the height above ground level that Google Earth™ will display the icon for the placemark;
- - a command that tells the earth visualization software where to find the icon image to use for each placemark. The icons used here are the flags of the country where the research organization to which the corresponding author belongs is located;
- - the year of publication of the citation to enable the “time slider” function.
Ancillary data:
Results and Discussion
Instructions for installation and use of the tropical marine microbial natural product geobibliography supplied with this article
- If not already done, install Google Earth™ free version from http://earth.google.com/
- Download file “Tropical marine microbial natural products geo bibliography.kml”
- Download file “Supporting KML files marine microbial natural products geobibliography.kmz” which is a compilation of the KML files containing the additional datasets listed in Table 1 and described in the ensuing text
- Open Google Earth™
- Use File>Open in the menu and open the abovementioned KML files
- A list of the datasets will appear in a window on the right hand side of the screen and the display of the each dataset, and the elements within, can be switched on and off by clicking on their checkboxes
- Note that some datasets require internet connections to interactively download required information and data download may take significant time. A small moving icon is often displayed by Google Earth™ to indicate that downloading is occurring.
References and Notes
- Abrell, LM; Cheng, X-C; Crews, P. New Nectriapyrones by Salt Water Culture of a Fungus Separated from an Indo-Pacific Sponge. Tetrahedron Letters 1994, 35, 9159–9160. [Google Scholar]
- Abrell, LM; Borgeson, B; Crews, P. A New Polyketide, Secocurvularin from the Salt Water Culture of a Sponge Derived Fungus. Tetrahedron Letters 1996, 37, 8983–8984. [Google Scholar]
- Adams, MWW; Perier, FB; Kelly, RM. Extremozymes: Expanding the Limits of Biocatalysis. Biotechnology 1995, 13, 662–668. [Google Scholar]
- Adinarayana, G; Venkateshan, M; Bapiraju, V; Sujatha, P; Premkumar, J; Ellaiah, P; Zeeck, A. Cytotoxic Compounds from the Marine Actinobacterium Streptomyces corchorusii Aubn1/71. Russian Journal of Bioorganic Chemistry 2006, 32, 295–300. [Google Scholar]
- Amagata, T; Rath, C; Rigot, JF; Tarlov, N; Tenney, K; Valeriote, FA; Crews, P. Structures and Cytotoxic Properties of Trichoverroids and Their Macrolide Analogues Produced by Saltwater Culture of Myrothecium verrucaria. J. Med. Chem 2003, 46, 4342–4350. [Google Scholar]
- Anonymous, Maritime Boundaries Geodatabase. In Vlaams Instituut voor de Zee: 2005.
- Asolkar, RN; Maskey, RP; Helmke, E; Laatsch, H. Chalcomycin B, a New Macrolide Antibiotic from the Marine Isolate Streptomyces Sp.B7064. J. Antibiot 2002, 55, 893–898. [Google Scholar]
- Bachrach, AJ. A Pictorial History of Diving; Best Publishing Company: San Pedro CA, 1988. [Google Scholar]
- Barsby, T; Kelly, MT; Gagne, SM; Andersen, RJ. Bogorol A Produced in Culture by a Marine Bacillus Sp. Reveals a Novel Template for Cationic Peptide Antibiotics. Org Lett 2001, 3, 437–440. [Google Scholar]
- Battershill, C; Jaspars, M; Long, P. Marine Biodiscovery: New Drugs from the Ocean Depths. Biologist 2005, 52, 107–114. [Google Scholar]
- Behrenfeld, MJ; Falkowski, PG. Photosynthetic Rates Derived from Satellite-Based Chlorophyll Concentration. Limnology and Oceanography 1997, 42, 1–20. [Google Scholar]
- Belofsky, GN; Jensen, PR; Renner, MK; Fenical, W. New Cytotoxic Sesquiterpenoid Nitrobenzoyl Esters from a Marine Isolate of the Fungus Aspergillus versicolor. Tetrahedron 1998, 54, 1715–1724. [Google Scholar]
- Belofsky, GN; Anguera, M; Jensen, PR; Fenical, W; Köck, M. Oxepinamides A-C and Fumiquinazolines H-I: Bioactive Metabolites from a Marine Isolate of a Fungus of the Genus Acremonium. Chem. Eur. J 2000, 6, 1355–1360. [Google Scholar]
- Berlinck, R; Hajdu, E; da Rocha, R; de Oliveira, J; Herna′ndez, I; Seleghim, M; Granato, A; de Almeida, E; Nunez, C; Muricy, G; Peixinho, S; Pessoa, C; Moraes, M; Cavalcanti, B; Nascimento, G; Thiemann, O; Silva, O; Souza, O; Silva, C; Minarini, P. Challenges and Rewards of Research in Marine Natural Products Chemistry in Brazil. J. Nat. Prod 2004, 67, 510–522. [Google Scholar]
- Boot, CM; Tenney, K; Valeriote, FA; Crews, P. Highly N-Methylated Linear Peptides Produced by an Atypical Sponge-Derived Acremonium Sp. J. Nat. Prod 2006, 69, 83– 92. [Google Scholar]
- Bugni, TS; Abbanat, D; Bernan, VS; Maiese, WM; Greenstein, M; Van Wagoner, RM; Ireland, CM. Yanuthones: Novel Metabolites from a Marine Isolate of Aspergillus niger. J. Org. Chem 2000, 6, 7195–7200. [Google Scholar]
- Bugni, TS; Janso, JE; Williamson, RT; Feng, X; Bernan, VS; Greenstein, M; Carter, GT; Maiese, WM; Ireland, CM. Dictyosphaeric Acids A and B: New Decalactones from an Undescribed Penicillium Sp. Obtained from the Alga Dictyosphaeria versluyii. J. Nat. Prod 2004, 67, 1396–1399. [Google Scholar]
- Byron, T. History of Spearfishing and Scuba Diving in Australia - the First 80 Years 1917 – 1997; Tom Byron Publishers: Sydney, Australia, 1998. [Google Scholar]
- Cheng, X; Jensen, P; Fenical, W. Luisols A and B, New Aromatic Tetraols Produced by an Estuarine Marine Bacterium of the Genus Streptomyces (Actinomycetales). J Nat Prod 1999, 62, 608–10. [Google Scholar]
- Connolly-Stone, K. The Interface with Existing Intellectual Property Systems: Limits and Opportunities for Existing Intellectual Property Rights; International Expert Workshop on Access to Genetic Resources and Benefit Sharing, Cuernavaca, Mexico, October 24–27, 2004; Bellot-Rojas, M, Bernier, S, Eds.; Cuernavaca: Mexico, 2004; pp. 147–152. [Google Scholar]
- Cueto, M; Jensen, P; Fenical, W. N-Methylsansalvamide, a Cytotoxic Cyclic Depsipeptide from a Marine Fungus of the Genus Fusarium. Phytochemistry 2000, 55, 223–6. [Google Scholar]
- Cueto, M; Jensen, PR; Kauffman, C; Fenical, W; Lobkovsky, E; Clardy, J. Pestalone, a New Antibiotic Produced by a Marine Fungus in Response to Bacterial Challenge. J. Nat. Prod 2001, 64, 1444–1446. [Google Scholar]
- Desjardine, K; Pereira, A; Wright, H; Matainaho, T; Kelly, M; Andersen, RJ. Tauramamide, a Lipopeptide Antibiotic Produced in Culture by Brevibacillus laterosporus Isolated from a Marine Habitat: Structure Elucidation and Synthesis. J Nat Prod 2007, 70, 1850–1853. [Google Scholar]
- Edwards, DJ; Marquez, BL; Nogle, LM; McPhail, K; Goeger, DE; Roberts, MA; Gerwick, WH. Structure and Biosynthesis of the Jamaicamides, New Mixed Polyketide-Peptide Neurotoxins from the Marine Cyanobacterium Lyngbya majuscule. Chemistry and Biology 2004, 11, 817–833. [Google Scholar]
- Ellis, E; Ramankutty, N. Putting People in the Map: Anthropogenic Biomes of the World. Frontiers in Ecology and the Environment, 2008. In Press.. [Google Scholar]
- Evans-Illidge, EA; Battershill, CN. Marine Natural Products Discovery in Australia: From Reef to Royalty, and the Pursuit of Convention for Biological Diversity (CBD) Compliance. Journal of Biolaw & Business Spec. Suppl 2007, 23–27. [Google Scholar]
- Feling, RH; Buchanan, GO; Mincer, TJ; Kauffman, CA; Jensen, PR; Fenical, W. Salinosporamide A: A Highly Cytotoxic Proteasome Inhibitor from a Novel Microbial Source, a Marine Bacterium of the New Genus Salinospora. Angew. Chem. Int. Ed 2003, 42, 355–357. [Google Scholar]
- Fenical, W. Chemical Studies of Marine Bacteria: Developing a New Resource. Chem Rev 1993, 93, 1673– 1683. [Google Scholar]
- Fernandez, JC. Elements for the Design of a Certificate of Legal Provenance; International Expert Workshop on Access to Genetic Resources and Benefit Sharing, Cuernavaca, Mexico, October 24–27, 2004; Bellot-Rojas, M, Bernier, S, Eds.; Cuernavaca: Mexico, 2004; pp. 267–270. [Google Scholar]
- Ford, PW; Gadepalli, M; Davidson, BS. Halawanones A-D, New Polycyclic Quinones from a Marine-Derived Streptomycete. J. Nat. Prod 1998, 61, 1232– 1236. [Google Scholar]
- Garo, E; Starks, CM; Jensen, PR; Fenical, W; Lobkovsky, E; Clardy, J. Trichodermamides A and B, Cytotoxic Modified Dipeptides from the Marine-Derived Fungus Trichoderma virens. J. Nat. Prod 2003, 66, 423–426. [Google Scholar]
- Gautschi, JT; Amagata, T; Amagata, A; Valeriote, FA; Mooberry, SL; Crews, P. Expanding the Strategies in Natural Product Studies of Marine-Derived Fungi: A Chemical Investigation of Penicillium Obtained from Deep Water Sediment. J Nat Prod 2004, 67, 362–367. [Google Scholar]
- Gerard, JM; Haden, P; Kelly, MT; Andersen, RJ. Loloatins A-D, Cyclic Decapeptide Antibiotics Produced in Culture by a Tropical Marine Bacterium. J. Nat. Prod 1999, 62, 80–85. [Google Scholar]
- Girsberger, MA. Disclosure of the Source of Genetic Resources and Traditional Knowledge in Patent Applications; International Expert Workshop on Access to Genetic Resources and Benefit Sharing, October 24–27, 2004; Bellot-Rojas, M, Bernier, S, Eds.; 2004; pp. 141–146. [Google Scholar]
- Gorajana, A; Kurada, BVVSN; Peela, S; Jangam, P; Vinjamuri, S; Poluri, E; Zeeck, A. 1-Hydroxy-1-Norresistomycin, a New Cytotoxic Compound from a Marine Actinomycete, Streptomyces chibaensis Aubn1/7. Journal of Antibiotics 2005, 58, 526–529. [Google Scholar]
- Guella, G; Dini, F; Erra, F; Pietra, F. Raikovenal, a New Sesquiterpenoid Favouring Adaptive Radiation of the Marine Ciliate Euplotes raikovi, and its Putative Biogenetic Precursor, Preraikovenal. J. Chem. Soc., Chem. Commun 1994, 1994, 2585– 2586. [Google Scholar]
- Guella, G; Pietra, F; Dini, F. Rarisetenolide, Epoxyrarisetenolide, and Epirarisetenolide, New-Skeleton Sesquiterpene Lactones as Taxonomic Markers and Defensive Agents of the Marine Ciliated Morphospecies, Euplotes rariseta. Helvetica Chimica Acta 2004, 79, 2180– 2189. [Google Scholar]
- Halpern, BS; Walbridge, S; Selkoe, KA; Kappel, CV; Micheli, F; D'Agrosa, C; Bruno, JF; Casey, KS; Ebert, C; Fox, HE; Fujita, R; Heinemann, D; Lenihan, HS; Madin, EMP; Perry, MT; Selig, ER; Spalding, M; Steneck, R; Watson, R. A Global Map of Human Impact on Marine Ecosystems. Science 2008, 319, 948– 952. [Google Scholar]
- Holler, U; Konig, GM; Wright, AD. Three New Metabolites from Marine-Derived Fungi of the Genera Coniothyrium and Microsphaeropsis. J. Nat. Prod 1999, 62, 114–118. [Google Scholar]
- Höller, U; König, GM; Wright, AD. A New Tyrosine Kinase Inhibitor from a Marine Isolate of Ulocladium botrytis and New Metabolites from the Marine Fungi Asteromyces cruciatus and Varicosporina ramulosa. Eur. J. Org. Chem 1999, 1999, 2949– 2955. [Google Scholar]
- Huang, H; She, Z; Lin, Y; Vrijmoed, LLP; Lin, W. Cyclic Peptides from an Endophytic Fungus Obtained from a Mangrove Leaf (Kandelia candel). J. Nat. Prod 2007, 70, 1696– 1699. [Google Scholar]
- IMF International Monetary Fund. World Economic Outlook Database. 2004. http://www.Imf.Org. [Google Scholar]
- Isaka, M; Suyarnsestakorn, C; Tanticharoen, M. Aigialomycins A-E, New Resorcylic Macrolides from the Marine Mangrove Fungus Aigialus parvus. J. Org. Chem 2002, 67, 1561–1566. [Google Scholar]
- Itoh, T; Kinoshita, M; Aoki, S; Kobayashi, M. Komodoquinone A, a Novel Neuritogenic Anthracycline, from Marine Streptomyces Sp. Ks3. J. Nat. Prod 2003, 66, 1373–1377. [Google Scholar]
- Jadulco, R; Proksch, P; Wray, V; Sudarsono; Berg, A; Grafe, U. New Macrolides and Furan Carboxylic Acid Derivative from the Sponge-Derived Fungus Cladosporium herbarum. J. Nat. Prod 2001, 64, 527– 530. [Google Scholar]
- Jadulco, R; Brauers, G; Edrada, RA; Ebel, R; Wray, V; Sudarsono; Proksch, P. New Metabolites from Sponge-Derived Fungi Curvularia lunata and Cladosporium herbarum. J. Nat. Prod 2002, 65, 730–733. [Google Scholar]
- Jaruchoktaweechai, C; Suwanborirux, K; Tanasupawatt, S; Kittakoop, P; Menasveta, P. New Macrolactins from a Marine Bacillus Sp. Sc026. J. Nat. Prod 2000, 63, 984–986. [Google Scholar]
- Jenkins, K; Jensen, P; Fenical, W. Thraustochytrosides A-C: New Glycosphingolipids from a Unique Marine Protist, Thraustochytrium globosum. Tetrahedron Lett 1999, 40, 7637–7640. [Google Scholar]
- Jenkins, KM; Renner, MK; Jensen, PR; Fenical, W. Exumolides A and B: Antimicroalgal Cyclic Depsipeptides Produced by a Marine Fungus of the Genus Scytalidium. Tetrahedron Letters 1998, 39, 2463–2466. [Google Scholar]
- Jeong, S-Y; Kim, HJSTS; Park, H-SLS-k; Kim, HM. Streptokordin, a New Cytotoxic Compound of the Methylpyridine Class from a Marine-Derived Streptomyces Sp. Kordi-3238. J. Antibiot 2006, 59, 234–240. [Google Scholar]
- Jiang, S; Sun, W; Chen, M; Dai, S; Zhang, L; Liu, Y; Lee, KJ; Li, X. Diversity of Culturable Actinobacteria Isolated from Marine Sponge Haliclona Sp. Antonie Van Leeuwenhoek 2007, 405–16. [Google Scholar]
- Kito, K; Ookura, R; Yoshida, S; Namikoshi, M; Ooi, T; Kusumi, T. Pentaketides Relating to Aspinonene and Dihydroaspyrone from a Marine-Derived Fungus, Aspergillus ostianus. J. Nat. Prod 2007, 70, 2022–2025. [Google Scholar]
- Kobayashi, J; Kubota, T. Bioactive Macrolides and Polyketides from Marine Dinoflagellates of the Genus Amphidinium. J. Nat. Prod 2007, 70, 451–460. [Google Scholar]
- Komatsu, K; Shigemori, H; Mikami, Y; Kobayashi, JI. Sculezonones A and B, Two Metabolites Possessing a Phenalenone Skeleton from a Marine-Derived Fungus Penicillium Species. J. Nat. Prod 2000, 63, 408–409. [Google Scholar]
- Krick, A; Kehraus, S; Gerhauser, C; Klimo, K; Nieger, M; Maier, A; Fiebig, H-H; Atodiresei, I; Raabe, G; Fleischhauer, J; Konig, GM. Potential Cancer Chemopreventive in Vitro Activities of Monomeric Xanthone Derivatives from the Marine Algicolous Fungus Monodictys putredinis. J. Nat. Prod 2007, 70, 353–360. [Google Scholar]
- Kubota, T; Tsuda, M; Takahashi, M; Ishibashi, M; Naoki, H; Kobayashi, J. Colopsinols B and C, New Long Chain Polyhydroxy Compounds from Cultured Marine Dinoflagellate Amphidinium Sp. J. Chem. Soc., Perkin Trans 1999, 1, 3483– 3487. [Google Scholar]
- Kumar, SS; Philip, R; Achuthankutty, CT. Antiviral Property of Marine Actinomycetes against White Spot Syndrome Virus in Penaeid Shrimps. Current Science 2006, 91, 807–811. [Google Scholar]
- Kwon, HC; Kauffman, CA; Jensen, PR; Fenical, W. Marinomycins A-D, Antitumor-Antibiotics of a New Structure Class from a Marine Actinomycete of the Recently Discovered Genus “Marinispora”. J. Am. Chem. Soc 2006, 128, 1622–1632. [Google Scholar]
- Laird, S; Monagle, C; Johnston, S. Queensland Biodiscovery Collaboration - the Griffith University Astra Zeneca Partnership for Natural Product Discovery - an Access and Benefit Sharing Case Study; United Nations University Institute of Advanced Studies, 2008; p. 54. [Google Scholar]
- Lin, W; Brauers, G; Ebel, R; Wray, V; Sudarsono, AB; Proksch, P. Novel Chromone Derivatives from the Fungus Aspergillus versicolor Isolated from the Marine Sponge Xestospongia exigua. J. Nat. Prod 2003, 66, 57–61. [Google Scholar]
- Lin, Y; Shao, Z; Jiang, G; Zhou, S; Cai, J; Vrijmoed, LLP; Gareth Jones, EB. Penicillazine, a Unique Quinolone Derivative with 4H-5,6-Dihydro-1,2-Oxazine Ring System from the Marine Fungus Penicillium Sp. (Strain #386) from the South China Sea. Tetrahedron 2000, 56, 9607–9609. [Google Scholar]
- Long, PF; Dunlap, WC; Battershill, CN; Jaspars, M. Shotgun Cloning and Heterologous Expression of the Patellamide Gene Cluster as a Strategy to Achieving Sustained Metabolite Production. ChemBioChem 2005, 6, 1760–1765. [Google Scholar]
- Magarvey, NA; Keller, JM; Bernan, V; Dworkin, M; Sherman, DH. Isolation and Characterization of Novel Marine-Derived Actinomycete Taxa Rich in Bioactive Metabolites. Applied and Environmental Microbiology 2004, 70, 7520–7529. [Google Scholar]
- Malmstrøm, J; Christophersen, C; Barrero, AF; Oltra, JE; Justicia, J; Rosales, A. Bioactive Metabolites from a Marine-Derived Strain of the Fungus Emericella variecolor. J. Nat. Prod 2002, 65, 364–367. [Google Scholar]
- Manning, TJ; Rhodes, E; Land, M; Parkman, R; Sumner, B; Lam, TT; Marshall, AG; Phillips, D. Impact of Environmental Conditions on the Marine Natural Product Bryostatin 1. Nat Prod Res 2006, 20, 611–28. [Google Scholar]
- McDonald, L; Barbieri, L; Bernan, V; Janso, J; Lassota, P; Carter, G. 07H239-A, a New Cytotoxic Eremophilane Sesquiterpene from the Marine-Derived Xylariaceous Fungus Ll-07h239. J. Nat. Prod 2004, 67, 1565–1567. [Google Scholar]
- McPhaden, MJ; Zebiak, SE; Glantz, MH. ENSO as an Integrating Concept in Earth Science. Science 2006, 314, 1740–1745. [Google Scholar]
- Miao, L; Kwong, T; Qian, P. Effect of Culture Conditions on Mycelial Growth, Antibacterial Activity, and Metabolite Profiles of the Marine-Derived Fungus Arthrinium C.F. saccharicola. Appl. Microbiol. Biotechnol 2006, 72, 1063–1073. [Google Scholar]
- Moore, BS; Trischman, JA; Seng, D; Kho, D; Jensen, PR; Fenical, W. Salinamides, Antiinflammatory Depsipeptides from a Marine Streptomycete. J. Org. Chem 1999, 64, 1145–1150. [Google Scholar]
- Motti, C; Bourne, DG; Burnell, JN; Doyle, JR; Haines, DS; Liptrot, CH; Llewellyn, LE; Ludke, S; Muirhead, A; Tapiolas, DM. Screening Marine Fungi for Inhibitors of the C4 Plant Enzyme Pyruvate Phosphate Dikinase: Unguinol as a Potential Novel Herbicide Candidate. Appl Environ Microbiol 2007, 73, 1921–7. [Google Scholar]
- Munro, MHG; Blunt, JW; Lake, RJ; Litaudon, M; Battershill, CN; Page, M. From Seabed to Sickbed: What Are the Prospects? “Sponges in time and space”.. Proc. 4th International Sponge Symposium, Amsterdam, April 19–23, 1993; van Soest, R, van Kempen, TMG, Braekman, J-C, Eds.; Amsterdam, 1993; pp. 473– 484.
- Murata, M; Legrand, AM; Ishibashi, Y; Fukui, M; Yasumoto, T. Structures and Configurations of Ciguatoxin from the Moray Eel Gymnothorax javanicus and Its Likely Precursor from the Dinoflagellate Gambierdiscus toxicus. J. Am. Chem Soc 1990, 112, 4380–4386. [Google Scholar]
- Nagai, H; Murata, M; Torigoe, K; Satake, M; Yasumoto, T. Gambieric Acids, New Potent Antifungal Substances with Unprecedented Polyether Structures from a Marine Dinoflagellate Gambierdiscus toxicus. J. Org. Chem 1992, 57, 5448–5453. [Google Scholar]
- Nakahara, T; Yokochi, T; Higashihara, T; Tanaka, S; Yaguchi, T; Honda, D. Production of Docosahexaenoic and Docosapentaenoic Acids by Schizochytrium sp Isolated from Yap Islands. J. Am. Oil Chem. Soc 1996, 73, 1421–1426. [Google Scholar]
- Namikoshi, M; Kobayashi, H; Yoshimoto, T; Meguro, S. Paecilospirone, a Unique Spiro[Chroman-2,1(3H)-Isobenzofuran] Derivative Isolated from Tropical Marine Fungus Paecilomyces sp. Chemistry Letters 2000, 29, 308. [Google Scholar]
- Namikoshi, M; Akano, K; Meguro, S; Kasuga, I; Mine, Y; Takahashi, T; Kobayashi, H. A New Macrocyclic Trichothecene, 12,13-Deoxyroridin E, Produced by the Marine-Derived Fungus Myrothecium roridum Collected in Palau. J. Nat. Prod 2001, 64, 396–398. [Google Scholar]
- Newman, DJ; Hill, RT. New Drugs from Marine Microbes: The Tide Is Turning. J Ind Microbiol Biotechnol 2006, 33, 539– 544. [Google Scholar]
- Newman, DJ; Cragg, GM. Natural Products as Sources of New Drugs over the Last 25 Years. J. Nat. Prod 2007, 70, 461–477. [Google Scholar]
- Oh, D; Jensen, P; Fenical, W. Zygosporamide, a Cytotoxic Cyclic Depsipeptide from the Marine-Derived Fungus Zygosporium masonii. Tetrahedron Lett 2006, 47, 8625–8628. [Google Scholar]
- Oh, D; Strangman, W; Kauffman, C; Jensen, P; Fenical, W. Thalassospiramides A and B, Immunosuppressive Peptides from the Marine Bacterium Thalassospira sp. Org. Lett 2007, 9, 1525–1528. [Google Scholar]
- Oleinikova, G; Shevchenko, L; Kuznetsova, T; Mikhalov, V. A Novel Fungicide of the Iturin Group, Obtained from a Marine Isolate of Bacillus subtilis. Isolation, Physico-Chemical and Biochemical Properties, Identification. Antibiot Khimioter 1995, 40, 19–21. [Google Scholar]
- Park, Y; Gunasekera, S; Lopez, J; McCarthy, P; Wright, A. Metabolites from the Marine-Derived Fungus Chromocleista sp. Isolated from a Deep-Water Sediment Sample Collected in the Gulf of Mexico. J. Nat. Prod 2006, 69, 580–584. [Google Scholar]
- Pathirana, C; Jensen, PR; Fenical, W. Marinone and Debromomarinone Antibiotic Sesquiterpenoid Naphthoquinones of a New Structure Class from a Marine Bacterium. Tetrahedron Letters 1992, 33, 7663–7666. [Google Scholar]
- Paul, VJ; Arthur, KE; Ritson-Williams, R; Ross, C; Sharp, K. Chemical Defenses: From Compounds to Communities. Biol Bull 2007, 213, 226–251. [Google Scholar]
- Poch, GK; James, B. Gloer, Helicascolides A and B: New Lactones from the Marine Fungus Helicascus kanaloanus. J. Nat. Prod 1989, 52, 257–260. [Google Scholar]
- Rahbæk, L; Christophersen, C; Frisvad, J; Bengaard, HS; Larsen, S; Rassingj, BR. Insulicolide A: A New Nitrobenzoyloxy-Substituted Sesquiterpene from the Marine Fungus Aspergillus insulicola. J. Nat. Prod 1997, 60, 811–813. [Google Scholar]
- Renner, MK; Jensen, PR; Fenical, W. Neomangicols: Structures and Absolute Stereochemistries of Unprecedented Halogenated Sesterterpenes from a Marine Fungus of the Genus Fusarium. J. Org. Chem 1998, 63, 8346– 8354. [Google Scholar]
- Renner, MK; Jensen, PR; Fenical, W. Mangicols: Structures and Biosynthesis of a New Class of Sesterterpene Polyols from a Marine Fungus of the Genus Fusarium. J. Org. Chem 2000, 65, 4843–4852. [Google Scholar]
- Saha, M; Jr., DG; Ghosh, D; Garai, D; Jaisankar, P; Sarkar, KK; Dutta, PK; Das, S; Jha, T; Mukherjee, J. Studies on the Production and Purification of an Antimicrobial Compound and Taxonomy of the Producer Isolated from the Marine Environment of the Sundarbans. Applied Microbiology and Biotechnology 2005, 66, 497–505. [Google Scholar]
- Saha, M; Jaisankar, P; Das, S; Sarkar, KK; Roy, S; Besra, SE; Vedasiromani, JR; Ghosh, D; Sana, B; Mukherjee, J. Production and Purification of a Bioactive Substance Inhibiting Multiple Drug Resistant Bacteria and Human Leukemia Cells from a Salt-Tolerant Marine Actinobacterium sp. Isolated from the Bay of Bengal. Biotechnology Letters 2006, 28, 1083–1088. [Google Scholar]
- Sayers, E. Pubchem: An Entrez Database of Small Molecules. NLM Tech Bull 2005, e2. [Google Scholar]
- Selvin, J; Joseph, S; Asha, KRT; Manjusha, WA; Sangeetha, VS; Jayaseema, DM; Antony, MC; Vinitha, AJD. Antibacterial Potential of Antagonistic Streptomyces sp. Isolated from Marine Sponge; Dendrilla nigra. FEMS Microbiology Ecology 2004, 50, 117–122. [Google Scholar]
- Shigemori, H; Tenma, M; Shimazaki, K; Kobayashi, J. Three New Metabolites from the Marine Yeast Aureobasidium pullulans. J. Nat. Prod 1998, 61, 696–698. [Google Scholar]
- Simmons, TL; Andrianasolo, E; McPhail, K; Flatt, P; Gerwick, WH. Marine Natural Products as Anti-Cancer Drugs. Mol Cancer Ther 2005, 4, 333– 342. [Google Scholar]
- Singh, IP; Milligan, KE; Gerwick, WH. Tanikolide, a Toxic and Antifungal Lactone from the Marine Cyanobacterium Lyngbya majuscule. J. Nat. Prod 1999, 62, 1333–1335. [Google Scholar]
- Smith, CJ; Abbanat, D; Bernan, VS; Maiese, WM; Greenstein, M; Tahir, JJA; Ireland, CM. Novel Polyketide Metabolites from a Species of Marine Fungi. J. Nat. Prod 2000, 63, 142–145. [Google Scholar]
- Soria-Mercado, IE; Prieto-Davo, A; Jensen, PR; Fenical, W. Antibiotic Terpenoid Chloro-Dihydroquinones from a New Marine Actinomycete. J. Nat. Prod 2005, 68, 904–910. [Google Scholar]
- Sperry, S; Samuels, GJ; Crews, P. Vertinoid Polyketides from the Saltwater Culture of the Fungus Trichoderma longibrachiatum Separated from a Haliclona Marine Sponge. J. Org. Chem 1998, 63, 10011–10014. [Google Scholar]
- Tan, LT; Cheng, XC; Jensen, PR; Fenical, W. Scytalidamides A and B, New Cytotoxic Cyclic Heptapeptides from a Marine Fungus of the Genus Scytalidium. J. Org. Chem 2003, 68, 8767–8773. [Google Scholar]
- Tapiolas, DM; Roman, M; Fenical, W; Stout, TJ; Clardy, J. Octalactins A and B: Cytotoxic Eight-Membered-Ring Lactones from a Marine Bacterium, Streptomyces sp. J. Am. Chem. Soc 1991, 113, 4682–4683. [Google Scholar]
- Taylor, MW; Hill, RT; Piel, J; Thacker, RW; Hentschel, U. Soaking It Up: The Complex Lives of Marine Sponges and Their Microbial Associates. ISME J 2007, 1, 187–90. [Google Scholar]
- ten Kate, K; Laird, SA. The Commercial Use of Biodiversity: Access to Genetic Resources and Benefit; Earthscan, 1999; p. 398. [Google Scholar]
- UN Millennium Project. Investing in Development: A Practical Plan to Achieve the Millennium Development Goals. Overview; United Nations: New York, 2005; p. 75. [Google Scholar]
- Varoglu, M; Corbett, TH; Valeriote, FA; Crews, P. Asperazine, a Selective Cytotoxic Alkaloid from a Sponge-Derived Culture of Aspergillus niger. J. Org. Chem 1997, 62, 7078–7079. [Google Scholar]
- Varoglu, M; Crews, P. Biosynthetically Diverse Compounds from a Saltwater Culture of Sponge-Derived Aspergillus niger. J. Nat. Prod 2000, 63, 41–43. [Google Scholar]
- Vogel, G. The Inner Lives of Sponges. Science 2008, 320, 1028–1030. [Google Scholar]
- Wu, J; Long, L; Song, Y; Zhang, S; Li, Q; Huang, J; Xiao, Z. A New Unsaturated Glycoglycerolipid from a Cultured Marine Dinoflagellate Amphidinium carterae. Chem. Pharm. Bull 2005, 53, 330–332. [Google Scholar]
- Xu, X; de Guzman, FS; Gloer, JB; Shearer, CA. Stachybotrins A and B: Novel Bioactive Metabolites from a Brackish Water Isolate of the Fungus Stachybotrys sp. J. Org. Chem 1992, 57, 6700–6703. [Google Scholar]
- Xu, Y; Miao, L; Li, X-C; Xiao, X; Qian, P-Y. Antibacterial and Antilarval Activity of Deep-Sea Bacteria from Sediments of the West Pacific Ocean. Biofouling 2007, 23, 131– 137. [Google Scholar]
- Young, TR. A Simple Solution: Using Certificates of Legal Provenance as a Workable Component of a Functional Regime on Abs; International Expert Workshop on Access to Genetic Resources and Benefit Sharing, Cuernavaca, Mexico, October 24–27, 2004; Bellot-Rojas, M, Bernier, S, Eds.; Cuernavaca: Mexico, 2004; pp. 284–286. [Google Scholar]
- Zheng, W; DeMattei, JA; Wu, J-P; Duan, JJ-W; Cook, LR; Oinuma, H; Kishi, Y. Complete Relative Stereochemistry of Maitotoxin. J. Am. Chem. Soc 1996, 118, 7946–7968. [Google Scholar]
- Zheng, Z; Zeng, W; Huang, Y; Yanga, Z; Li, J; Cai, H; Su, W. Detection of Antitumor and Antimicrobial Activities in Marine Organism Associated Actinomycetes Isolated from the Taiwan Strait, China. FEMS Microbiology Letters 2000, 188, 87–91. [Google Scholar]













| Downloaded Data Sets (KML format unless otherwise indicated) | Source |
|---|---|
| Boundaries of Exclusive Economic Zones | Vlaams Instituut voor de Zee [6] from http://bbs.keyhole.com/ubb/showthreaded.php/Cat/0/Number/693959/page/vc/vc/1 |
| Bathymetric imagery | National Geophysical Data Centre, NOAA |
| Millennium Development Goals | Millennium Development Goal Monitor ( www.mdgmonitor.org) gives global socioeconomic information |
| Sea Surface Temperature composites for the period 1 December 2007 – 1 January 2008. | NASA Earth Observatory program ( www.neo.sci.gsfc.nasa.gov) |
| Chlorophyll composites for the period 1 December 2007 – 1 January 2008. | NASA Earth Observatory program, data from AQUA and MODIS satellites Available at www.neo.sci.gsfc.nasa.gov |
| Global population data | Center for International Earth Science Information Network and Centro Internacional de Agricultura Tropical, Colombia University, NY, USA. Google Earth™ version downloadable from www.bbs.keyhole.com/ubb/showflat.php/Cat/0/Number/92018/Main/90871 |
| Anthropogenic biomes. | Described by Ellis and Ramankutty [25] with KML file at www.eoearth.org/article/Anthropogenic_biome_maps |
| Impact of human activities | Google Earth™ file supporting Halpern et al [38] downloadable from www.nceas.ucsb.edu/GlobalMarine |
| Location of coral reefs. This data needed tobe converted to KML format. | Worldfish Center’s Reefbase ( www.reefbase.org) |
| Location of undersea volcanoes. | Smithsonian Institution Global Volcanism Program. Available for download at www.volcano.si.edu |
Share and Cite
Mukherjee, J.; Llewellyn, L.E.; Evans-Illidge, E.A. A Tropical Marine Microbial Natural Products Geobibliography as an Example of Desktop Exploration of Current Research Using Web Visualisation Tools. Mar. Drugs 2008, 6, 550-577. https://doi.org/10.3390/md20080028
Mukherjee J, Llewellyn LE, Evans-Illidge EA. A Tropical Marine Microbial Natural Products Geobibliography as an Example of Desktop Exploration of Current Research Using Web Visualisation Tools. Marine Drugs. 2008; 6(4):550-577. https://doi.org/10.3390/md20080028
Chicago/Turabian StyleMukherjee, Joydeep, Lyndon E. Llewellyn, and Elizabeth A. Evans-Illidge. 2008. "A Tropical Marine Microbial Natural Products Geobibliography as an Example of Desktop Exploration of Current Research Using Web Visualisation Tools" Marine Drugs 6, no. 4: 550-577. https://doi.org/10.3390/md20080028
APA StyleMukherjee, J., Llewellyn, L. E., & Evans-Illidge, E. A. (2008). A Tropical Marine Microbial Natural Products Geobibliography as an Example of Desktop Exploration of Current Research Using Web Visualisation Tools. Marine Drugs, 6(4), 550-577. https://doi.org/10.3390/md20080028