Acetylcholinesterase-Inhibiting Activity of Pyrrole Derivatives from a Novel Marine Gliding Bacterium, Rapidithrix thailandica
Abstract
:1. Introduction
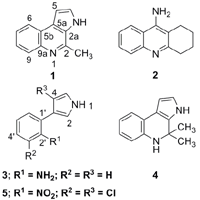
2. Results and Discussion

3. Experimental
3.1. General
3.2. Bacterial isolation and purification
3.3. Taxonomic identification
3.4. Large scale fermentation
3.5. Extraction and compound isolation
3.6. Bioactivity determination
Acknowledgments
References and Notes
- Racchi, M; Mazzuucchelli, M; Porrello, E; Lanni, C; Govoni, S. Acetylcholinesterase Inhibitors: Novel Activities of Old Molecules. Pharmacol. Res 2004, 50, 441–451. [Google Scholar]
- Lanari, A; Amenta, F; Silvestrelli, G; Tomassoni, D; Parnetti, L. Neurotransmitter Deficits in Behavioral and Psychological Symptoms of Alzheimer’s Disease. Mech. Ageing Dev 2006, 127, 158–165. [Google Scholar]
- Srisukchayakul, P; Suwannachart, C; Sangnoi, Y; Kanjanaopas, A; Hosoya, S; Yokota, A; Arunpairojana, V. Rapidithrix thailandica gen. nov., sp. nov., A Marine Gliding Bacteria Isolated from Andaman Sea, the Southern Coastline of Thailand. Int. J. Syst. Evol. Microbiol 2007, 54, 2275–2279. [Google Scholar]
- Kanjanaopas, A; Panphon, S; Fun, H-K; Chantrapromma, S. 4-Methyl-3H-pyrrolo[2,3-c]quinoline. Acta Cryst. E 2006, 62, o2728–o2730. [Google Scholar]
- Chang, CJ; Floss, HG; Hook, DJ; Mabe, JA; Manni, PE; Martin, LL; Schröder, K; Shieh, TL. The Biosynthesis of the Antibiotic Pyrrolnitrin by Pseudomonas aureofaciens. J. Antibiot 1981, 34, 555–566. [Google Scholar]
- van Pée, K-H; Salcher, O; Fischer, P; Bokel, M; Lingens, F. The Biosynthesis of Brominated Pyrrolnitrin Derivatives by Pseudomonas aureofaciens. J. Antibiot 1983, 36, 1735–1742. [Google Scholar]
- Zhou, P; Mocek, U; Siesel, B; Floss, HG. Biosynthesis of Pyrrolnitrin. Incorporation of 13 C,15N Double-Labelled D- and L-Tryptophan. J. Basic Microbiol 1992, 32, 209–214. [Google Scholar]
- Gerth, K; Trowitzsch, W; Wray, V; Höfle, G; Irschik, H; Reichenbach, H. Pyrrolnitrin from Myxococcus fulvus (Myxobacterales). J. Antibiot 1982, 35, 1101–1103. [Google Scholar]
- El-Banna, N; Winkelmann, G. Pyrrolnitrin from Burkholderia capacia: Antibiotic Activity against Fungi and Novel Activities against Streptomycetes. J. Appl. Microbiol 1998, 85, 69–78. [Google Scholar]
- Pohanka, A; Broberg, A; Johansson, M; Kenne, L; Lavenfors, J. Pseudotrienic Acids A and B, Two Bioactive Metabolites from Pseudomonas sp. MK381-IODS. J. Nat. Prod 2005, 68, 1380–1385. [Google Scholar]
- Kirner, S; Hammer, PE; Hill, DS; Altmann, A; Fischer, I; Weislo, LJ; Lanahan, M; van Pée, K-H; Ligon, JM. Functions Encoded by Pyrrolnitrin Biosynthesis Genes from Pseudomonas fluorescens. J. Bacteriol 1998, 180, 1939–1943. [Google Scholar]
- Muñoz-Ruiz, P; Rubio, L; García-Palomero, E; Dorrensoro, I; del Monte-Millán, M; Valenzuela, R; Usán, P; de Austria, C; Bartolini, M; Andrisano, V; Bidon-Chanal, A; Orozco, M; Luque, FJ; Medina, M; Martínez, A. Design, Synthesis, and Biological Evaluation of Dual Binding Site Acetylcholinesterase Inhibitors: New Disease-Modifying Agents for Alzheimer’s Disease. J. Med. Chem 2005, 48, 7223–7233. [Google Scholar]
- da Silva, CHTP; Campo, VL; Carvalho, I; Taft, CA. Molecular Modeling, Docking and ADMET Studies Applied to the Design of a Novel Hybrid for Treatment of Alzheimer’s Disease. J. Mol. Graph. Model 2006, 25, 169–175. [Google Scholar]
- Hosoya, S; Arunpairojana, V; Suwannachart, C; Kanjanaopas, A; Yokota, A. Aureispira marina gen. nov., sp., nov., a Gliding Arachidonic Acid-Containing Bacterium Isolated from the Southern Coastline of Thailand. Int. J. Syst. Evol. Microbiol 2006, 56, 2931–2935. [Google Scholar]
- Zhang, PL; Wang, YH; Fang, MX; Stackebrandt, E; Ding, BY. Improved Methods of Isolation and Purification of Myxobacteria and Development of Fruiting Body Formation of Two Strains. J. Microbiol. Meth 2003, 54, 21–27. [Google Scholar]
- Reichenbach, H. The Prokaryotes; Balows, A, Truper, HG, Dworkin, M, Harder, W, Schleifer, KH, Eds.; Springer-Verlog: New York, 1992; Vol. 4, The Order Cytophagales; pp. 3631–3675. [Google Scholar]
- Hall, TA. Bioedit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Window 95/98/NT. Nucleic Acids Symp. Ser 1999, 41, 765–768. [Google Scholar]
- Suzuki, Y; Ojia, M; Sakagami, Y; Fudou, R; Yamanaka, S. Cystothiazoles C-F, New Bithiazole-Type Antibiotics from the Myxobacterium Cystobacter fuscus. Tetrahedron 1998, 54, 11399–11404. [Google Scholar]
- Vobis, G. The Prokaryotes; Balows, A, Truper, HG, Dworkin, M, Harder, W, Schleifer, KH, Eds.; Springer-Verlog: New York, 1992; Vol. 2, The Genus Actinoplanes and Related Genera; pp. 1031–1060. [Google Scholar]
- Ingkaninan, K; Changwijit, K; Suwanborirux, K. Vobasinyl-Iboga Bisindole Alkaloids, Potent Acetylcholinesterase Inhibitors from Tabernaemontana divaricata Root. J. Pharm. Pharmacol 2006, 58, 847–852. [Google Scholar]
- Ellman, GL; Courtney, KD; Andres, V, Jr; Featherstone, RM. A New and Rapid Colorimetric Determination of Acetylcholinesterase Activity. Biochem. Pharmacol 1961, 7, 88–95. [Google Scholar]
- Skehan, P; Storeng, R; Scudiero, D; Monks, A; McMahan, J; Vistica, D; Warren, JT; Bokesch, H; Kenney, S. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst 1990, 82, 1107–1112. [Google Scholar]
| position | 1Ha (mult.; J in Hz) | 13C (mult.) |
|---|---|---|
| 1 | 10.90 (br s) | - |
| 2 | 6.96 (ddd; 2.3,2.3,<1) | 115.8 (CH) |
| 3 | - | 121.1 (C) |
| 4 | 6.25 (ddd; 2.5,2.3,<1) | 107.3 (CH) |
| 5 | 6.83 (ddd; 2.5,2.3,2.3) | 118.3 (CH) |
| 1′ | - | 121.6 (C) |
| 2′ | - | 144.8 (C) |
| 3′ | 6.69 (dd; 7.9,1.3) | 115.2 (CH) |
| 4′ | 6.88 (ddd; 7.9,7.3,1.5) | 126.2 (CH) |
| 5′ | 6.55 (ddd; 7.5,7.3,1.3) | 116.9 (CH) |
| 6′ | 7.10 (dd; 7.5,1.5) | 128.9 (CH) |
| 2′-NH2 | 4.20 (br s; 2H) | - |
| position | 1Ha (mult.; J in Hz) | 13Cb (mult.) |
|---|---|---|
| 1 | 5.70 (br s) | - |
| 2 | - | 52.1 (C) |
| 2a | - | 132.2 (C) |
| 3 | 10.74 (br s) | - |
| 4 | 6.64 (dd; 2.8,2.6) | 117.8 (CH) |
| 5 | 6.20 (dd; 2.6,2.4) | 101.5 (CH) |
| 5a | - | 113.5 (C) |
| 5b | - | 119.1 (C) |
| 6 | 7.08 (dd; 7.5,1.3) | 121.0 (CH) |
| 7 | 6.45 (ddd; 7.7,7.5,1.0) | 116.2 (CH) |
| 8 | 6.73 (ddd; 7.9,7.7,1.3) | 125.0 (CH) |
| 9 | 6.50 (dd; 7.9,1.0) | 112.7 (CH) |
| 9a | - | 141.7 (C) |
| 2-CH3 | 1.42 (s; 6H) | 30.6 (CH3, 2C) |
Share and Cite
Sangnoi, Y.; Sakulkeo, O.; Yuenyongsawad, S.; Kanjana-opas, A.; Ingkaninan, K.; Plubrukarn, A.; Suwanborirux, K. Acetylcholinesterase-Inhibiting Activity of Pyrrole Derivatives from a Novel Marine Gliding Bacterium, Rapidithrix thailandica. Mar. Drugs 2008, 6, 578-586. https://doi.org/10.3390/md6040578
Sangnoi Y, Sakulkeo O, Yuenyongsawad S, Kanjana-opas A, Ingkaninan K, Plubrukarn A, Suwanborirux K. Acetylcholinesterase-Inhibiting Activity of Pyrrole Derivatives from a Novel Marine Gliding Bacterium, Rapidithrix thailandica. Marine Drugs. 2008; 6(4):578-586. https://doi.org/10.3390/md6040578
Chicago/Turabian StyleSangnoi, Yutthapong, Oraphan Sakulkeo, Supreeya Yuenyongsawad, Akkharawit Kanjana-opas, Kornkanok Ingkaninan, Anuchit Plubrukarn, and Khanit Suwanborirux. 2008. "Acetylcholinesterase-Inhibiting Activity of Pyrrole Derivatives from a Novel Marine Gliding Bacterium, Rapidithrix thailandica" Marine Drugs 6, no. 4: 578-586. https://doi.org/10.3390/md6040578
APA StyleSangnoi, Y., Sakulkeo, O., Yuenyongsawad, S., Kanjana-opas, A., Ingkaninan, K., Plubrukarn, A., & Suwanborirux, K. (2008). Acetylcholinesterase-Inhibiting Activity of Pyrrole Derivatives from a Novel Marine Gliding Bacterium, Rapidithrix thailandica. Marine Drugs, 6(4), 578-586. https://doi.org/10.3390/md6040578