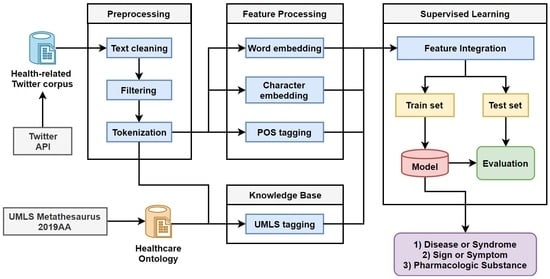
1. Introduction
An overwhelming amount of health-related knowledge has been recorded in social media sites such as Twitter, with the number of tweets posted each year increasing exponentially [
1,
2,
3]. Twitter is the most comprehensive social media site collecting and providing public health information: 500 million tweets are sent each day—5000 every second. Although a large amount of information is thought to be reliable for monitoring and analyzing health-related information, the lack of methodological transparency for data extraction, processing, and analysis has led to inaccurate predictions in detecting disease outbreaks, adverse drug events, etc. As a result, health-related text mining and information extraction are active challenges for the development of useful public health applications for researchers [
4,
5,
6]. One essential part of developing such an information extraction system is the NER process, which defines the boundaries between common words in terminology in a particular text, and assigns the terminology to specific categories based on domain knowledge [
7,
8,
9].
NER, also known as entity extraction, classifies named entities that are present in a text into pre-defined categories like “location”, “time”, “person”, “organization”, “money”, “percent”, and “date”, etc. [
10]. An example is as follows: (ORG U.N.) official (PER Ekeus) heads for (LOC Baghdad) [
11]. This sentence contains three named entities: Ekeus is a person, the U.N. is an organization, and Baghdad is a location.
In the traditional NER method based on machine learning, part-of-speech (POS) information is considered as a key feature of entity recognition [
10,
11,
12,
13]. In 2016, Lample et al. [
7] presented a neural architecture based on long short-term memory (LSTM) that uses no language-specific resources and hand-engineered features. They compared the LSTM and conditional random fields (LSTM-CRF) model and stack LSTM (S-LSTM) model with various NER tasks. The state-of-the-art NER systems for English produce near-human performance with an F1 score of over 90%. For example, the best system entering Seventh Message Understanding Conference (MUC-7) in [
14] scored 93.39% for the F-measure, while human annotators scored 97.60% and 96.95%. However, the performances in the healthcare, biomedical, chemical, and clinical domains are not as good as the performances in the English domain. They are restricted by problems such as the number of new terms being created on a regular basis, the lack of standardization of technical terms between authors, and by the fact that technical terms (for example, disease, drugs, and symptoms) often have multiple names [
15]. Consequently, state-of-the-art NER software (e.g., Stanford NER) is less effective on Twitter NER tasks [
9].
Public health research requires the knowledge of disease, drugs, and symptoms. Researchers focus on exploring population health, well-being, disability, and the determining factors for these statuses, be they biological, behavioral, social, or environmental. Moreover, researchers develop and assess interventions aiming to improve population health, prevent disease, compensate for disabilities, and provide innovations in terms of the organization of health, social, and medical services [
16]. The Internet has revolutionized efficient health-related communication and epidemic intelligence [
17]. People are increasingly using the Internet and social media channels. In the modern world of social media dominance, microblogs like Twitter are probably the best source of up-to-date information. Twitter provides a huge amount of microblogs, including health information that are completely public and pullable.
The purpose of the research reported in this paper was to predict health-related named entities such as diseases, symptoms, and pharmacologic substances from noisy Twitter messages that are essential for discovering public health information and developing real-time prediction systems with respect to disease outbreak prediction and drug interactions. To achieve this goal, we employed a deep learning approach obtaining the pre-trained word embedding which can be used successfully for any text mining tasks. We collected a large number of Twitter data, and then cleaned and preprocessed them to produce an experimental dataset. We automatically annotated the dataset using the UMLS Metathesaurus [
18] with three types of entities (diseases, symptoms, and pharmacologic substance). Our deep learning architecture follows the window approach in [
19]. The method we put forward has a number of desirable advantages:
We achieved a precision of 93.99%, recall of 73.31%, and F1-score of 81.77% for disease or syndrome HNER; a precision of 90.83%, recall of 81.98%, and F1-score of 87.52% for sign or symptom HNER; and a precision of 94.85%, recall of 73.47%, and F1-score of 84.51% for pharmacologic substance named entities using the BiLSTM-CRF model.
The architecture uses little hand-engineered features using POS tagging. Therefore, it has a great capability for improving state-of-the-art performances.
We presented a large number of tweets on the HNER task using domain-specific UMLS ontology, including three health-related entity types (diseases, symptoms, and pharmacologic substance).
The health-related domain (including disease, syndrome, sign, symptom, and pharmacologic substance) was particularly well applied because the BiLSTM-CRF could extract health-related entities and identify the relationship between them from Twitter messages.
The remainder of the paper is organized as follows:
Section 2 introduces the theoretical foundation of this paper and related works.
Section 3 focuses on the detailed description of the experimental dataset, health-related named entity recognition tasks, and how the deep learning model is trained. In
Section 4, the experimental analysis and the related results are provided. Finally,
Section 5 provides a discussion about the experimental analysis and address our conclusion.
3. Materials and Methods
3.1. Dataset
We have obtained a large number of health-related twitter data through Twitter API [
52] using the search term “healthcare” between 12 July 2018 and 12 July 2019. The dataset contains 1,403,393 health-related tweets.
For the HNER task, we only considered the three types of entities such as diseases, symptoms, and pharmacologic substances to match the particular entities we target for annotation. These types of entities are also annotated in Micromed dataset [
49].
Table 1 shows the detail of each entity type. We found 189,517 tweets for “disease or syndrome”, containing 382,629 medical terms (7.25% of total words) and 9536 unique terms (3.74% of total unique words). There were 77,466 tweets found for “sign or symptom”, containing 99,367 medical terms (4.33% of total words) and 2043 unique terms (4.56% of total unique words). A total of 409,268 tweets were found for “pharmacologic substance”, containing 848,871 medical terms (7.51% of total words) and 8148 unique terms (1.80% of total unique words). Examples of tweets and corresponding medical terms are as shown below:
Example 1: “Cannabis (T121) Strains (T121) to beat stress (T184) after recommendations from Marijuana (T121) doctors in Los Angeles”.
Example 2: “Join VLAB on February 26th to learn more about the breakthroughs in diabetes (T047) like the artificial pancreas (T047)”.
Example 3: “Nightmare (T184), narcolepsy (T184) and sudden (T184) weakness (T184) turn Mary’s life upside down after swine flu (T047) vaccination”.
In the preprocessing step, we removed all URLs (starting with “http” and “https”), hashtags (starting with “#”), non-English characters, and punctuation. Then we converted all characters to lower case. Finally, we only selected the tweets containing at least five words. Not all tweets contained health-related entities. We filtered out tweets using a list of medical terms in UMLS. We only kept the tweets if it contained at least one entity from the medical entity types, and the others were removed.
Finally, we filtered 676,251 tweets with a total of 1,330,867 medical terms and 19,727 unique medical terms for our experiment. The tweets in the experimental dataset contain at least one health-related entity. The health-related entities in each entity type and frequency are shown in
Table 2. To avoid a large number of false positives, we removed the following non-medical terms from each entity type:
- -
T047: condition, best, recruitment, disease, may, said, founder, increasing, west, evaluable, etc.
- -
T184: fit, weight, finding, catch, imbalance, medicine, others, walking, spots, mass, etc.
- -
T121: water, various, program, drugs, stop, tomorrow, orange, support, solution, speed, etc.
We joined the relevant Metathesaurus table (“MRCONSO.RRF” and “MRSTY.RRF”) to determine health-related named entities. We normalized all terms in tweets using the Jaccard similarity measure (>0.7):
- -
T047: diabet to diabeta (0.80), alzheime to alzheimer (0.86), obesit to obesity (0.80), etc.
- -
T184: strains to strain (0.80), grimaced to grimace (0.83), illnesss to illness (0.83), etc.
- -
T121: marijuan to marijuana (0.86), pharmaceutica to pharmaceutical (0.71), etc.
After all, preprocessing and filtering, we split the experimental dataset into training, testing, and validation subsets.
Table 3 shows the distribution of tweets and the corresponding number of tweets, number of terms, and unique terms for each entity type.
3.2. Dataset Annotation Tool
For dataset annotation, we used QuickUMLS tool [
53] to extract biomedical concepts from medical text. We use downloaded the latest version of UMLS (umls-2019AA-metathesaurus) and set the parameters as shown in
Table 4.
3.3. Health-Related Named Entity Recognition
In this section, we provide the problem definition in HNER, the details of BiLSTM-CRF model architecture and the process of the training. We apply the Pytorch library [
54] to implement our model. Our main goal is to predict medical terms in given sentences or tweets. The overview of BiLSTM-CRF model is shown in
Figure 2. BiLSTM-CRF model consists of four layers including the embedding, BiLSTM, CRF, and Viterbi layers. The embedding layer consists of the three sub representations such as word embedding features (yellow), character features (red), and additional word features (green). The medical and non-medical pre-trained word embeddings are used and compared for producing word embedding. CNN is used for producing character embedding, and POS tagging is used for producing additional word features. BiLSTM learns the contextual information from the concatenated word and character representations, and generates the word-level contextual representations that indicate the confidence score “CS” for each word. The CRF layer calculates tagging scores for each word input based on the contextual information. Finally, the Viterbi algorithm is used to find the tag sequence that maximizes the tagging scores. We explain the details of the presented model in the next sections and how it applies to the HNER task.
3.3.1. Problem Definition
We consider named entity recognition as a combination of two problems: segmentation and sequence labelling, given
- -
an ordered set of character sequences , where is a character sequence;
- -
an ordered set of annotations , where is a sequence and is a tuple of two boolean labels () showing whether the corresponding character is the beginning of a chemical entity and/or part of one, respectively.
Our task is to create a predictor , where is a set of inferred annotations similar to . We also use a tokenizer: , where is an ordered sequence of character subsequences (tokens), thus slightly redefining the objective function to target per-token annotations. Provided that the tokenizer is fine enough to avoid tokens with overlapping annotations, this redefined problem is equivalent to the original one.
3.3.2. Feature Representation
In the first phase of the prediction model, named as embedding, we represent each token by word embedding (1), character embedding (2), and POS tagging (3).
Word Embedding (word): We used both non-biomedical and biomedical pre-trained word embedding and analyzed the effect of word embedding for the HNER task. In this paper, we used non-medical word embedding with GloVe [
55] and Word2Vec [
56]. We also used medical word embedding as found in Pyyssalo et al. [
57], Chiu et al. [
47], Chen et al. [
58], and Aueb et al. [
59]. Our experimental results show the comparison of these word embedding on the healthcare NER task from Twitter. The details are explained in
Appendix A and the statistics of word embedding are described in
Table A1 and
Table A2.
Character Embedding (char): Character-level word embedding is useful, especially when rich rare words and out-of-vocabulary words are exploited and word embedding is poorly trained. It is common in the biomedical and chemical domain. Word-level approaches fall short when applied to Twitter data, where many infrequent or misspelled words occur within very short documents. We considered character-level word embedding in this paper. The details are explained in
Appendix B and. Also,
Table A3 shows the character set used in this paper and
Figure A1 shows the CNN for extracting character-level features.
Additional word feature (POS): Most state-of-the-art NER systems [
39,
60] use additional features such as POS tagging [
61] as a form of external knowledge. We also used POS tagging as an additional word feature in this paper. POS tags are useful for building parse trees, which are used in building NERs and extracting relations between words.
Table 5 shows an example of how POS features are applied.
3.3.3. Feature Learning
After concatenating the different feature representations, we employed the BiLSTM layer to learn sequential structure of words in tweets. LSTM and BiLSTM have commonly used RNN techniques in NLP tasks. In comparison with a single-direction LSTM, a BiLSTM can use the information from both sides to learn the input features. The details are explained in
Appendix C and
Figure A2 shows the LSTM memory cell in detail.
3.3.4. Prediction
After learning the input features, the famous CRF layer is employed. BiLSTM-CRF is the combination between BiLSTM and CRF, a string algorithm for sequence labelling tasks which is very effective. In a BiLSTM model, the tagging decision at the output layer is made independently using a softmax activation function. That means the final tagging decision of a token does not depend on the tagging decision of others. Therefore, adding a CRF layer into a BiLSTM model equips the model with the ability to learn the best sequence of tags that maximizes the log probability of the output tag sequence. BiLSTM-CRF is very successful for NER tasks. They produce the state-of-the-art results on several NER benchmark data sets without using any features. The details are explained in
Appendix D and
Appendix E.
3.4. Network Training
In this section, we provide the detail process of our neural network training. We apply the Pytorch library to implement the LSTM-CRF and BiLSTM-CRF models.
We train our network architecture with the back-propagation algorithm [
62] to update the parameters for each training example using the work of Adam [
63] with Nesterov momentum [
64]. In each epoch, we divide all the training data into batches, then process one batch at a time. The batch size decides the number of sentences. In each batch, we firstly get the output scores from the BiLSTM for all labels. Then we put the output scores into CRF layer, and we can get the gradient of outputs and the state transition matrix. From this, we can backpropagate the error from output to input, which contains the backward propagation for bi-directional states of LSTM. Finally, we update all the parameters.
Dropout [
65] can mitigate the overfitting problem. We apply dropout on the weight vectors directly to mask the final embedding layer before the combinational embedding feed into the bi-directional LSTM. We fix the dropout rate at 0.5 as usual and achieve good performance on our model. We also use the early stopping strategy with patience 20 to avoid overfitting the early stopping monitored weighted F1-scores on validation sets.
3.5. Hyparameter Settings
Our hyper-parameters are shown in
Table 6. We used three-layer convolution and set the output of the convolution layer to 50 for extracting character features from each word. We also used two-layer LSTM and set the state size of LSTM to 250. For stopping condition, we used an early stopping strategy, and maximum iteration has been set at 100. The batch size is 100, the dropout layer is 0.5, and the initial learning rate is 0.001.
The experimental hardware platform was the Intel Xeon E3 (32G memory, GTX 1080 Ti). The experimental software platform was the Ubuntu 17.10 operating system and the development environment was the Python 3.5 programming language. The Pytorch library and the Scikit-learn library of Python were used to build the healthcare NER recognition model and comparative experiments.
3.6. Evaluation Metrics
For evaluating our model, an exact matching criterion was used to examine three different result types. False-negative (FN) and false-positives (FP) are incorrect negative and positive predictions, respectively. True-positive (TP) results corresponded to correct positive predictions, which are actual correct predictions. The evaluation is based on the performance measures precision (P), recall (R), and F-score (F). Recall denotes the percentage of correctly labelled positive results overall positive cases and is calculated as:
4. Results and Discussion
In this paper, we employed the BiLSTM-CRF model with different combinations of word features (word embedding, character embedding, and POS tagging) for the divided dataset. The BilSTM-CRF model is compared with LSTM-CRF model presented by Jimeno-Yepes and MacKinlay [
50] for the most similar task. To best of our knowledge, there are no other published works which use Twitter data for the health-related NER task. They used LSTM-CRF model with a pre-trained word-embedding and outperformed CRF model on the Micromed dataset. We present a dataset similar to Micromed, but our dataset is larger. Larger datasets support deep learning methods to improve the complexity of the problem and of the learning algorithm. The comparative performance evaluation result is shown in
Table 7. The disease or syndrome HNER performance of BiLSTM-CRF (word + char + POS) has a precision of 93.99%, recall of 73.31%, and F1 of 81.77% when evaluating on the presented dataset. BiLSTM-CRF (word + char) has a precision of 94.53%, and LSTM-CRF (word + char + POS) has an F1 of 82.08%. The sign or symptom HNER performance of BiLSTM-CRF (word + char + POS) has a precision of 90.83%, recall of 81.98%, and F1 of 87.52%. The pharmacologic substance HNER performance of BiLSTM-CRF (word + char + POS) has a precision of 94.85%, recall of 73.47%, and F1 of 84.51%. BiLSTM-CRF (word + char) has a precision of 94.93%. Experimental results on the presented dataset show that BiLSTM-CRF (word + char + POS) could yield excellent performance for the HNER task. Surprisingly, the precision of BiLSTM-CRF without the POS tagging model for disease or syndrome is 0.54% higher, and for pharmacologic substance it is 0.08% higher than that of the BiLSTM-CRF with the POS tagging model when evaluating the presented dataset. Also, the F1 of LSTM-CRF with the all-features model for disease or syndrome is 0.31% higher than the BiLSTM-CRF with the-features model.
For these experiments, we used “Pyysalo Wiki + PM + PMC” word embeddings that achieve higher results than other pre-trained word embeddings (see
Table 8). As compared to the Micromed dataset and the presented dataset, the LSTM + CRF (word) model applied to both datasets. The model on the presented dataset improved the performance significantly. LSTM+CRF (word) model performed better results than LSTM + CRF (char) and LSTM + CRF (POS) models. We can see that word embedding is most effective feature for HNER task compared with character embedding and POS tagging. The models with different combinations of features improve the result. The best results are shown with BiLSTM-CRF (word + char + POS), using the combination of all feature types. The Twitter dataset is highly noisy and many out-of-vocabulary words are contained. Because of that, character embedding helps to learn more those words and other rare words. As we mentioned above, most of the state-of-the-art results used POS tagging. Also, our experimental result proves that POS tagging is efficient in various NER tasks. Generally, the BiLSTM + CRF model outperforms the LSTM + CRF model in all the experiments.
As shown in
Table 7, pre-trained word embedding is the most significant feature and can be used efficiently for down-stream tasks such as NER and HNER tasks. We achieved the best result with BiLSTM-CRF (word + char + POS) model. We studied the contribution of medical and non-medical word embeddings to BiLSTM-CRF (word +char + POS) model performance by removing each of them in turn from the model and then evaluating the model on the presented dataset. In this regard, we evaluate the model with character embedding and POS tagging.
Table 8 shows the predictive performance for the model with different word embeddings on the testing set. Generally, the models with non-medical pre-trained word embeddings achieve a higher result than medical pre-trained word embeddings. The experimental results show that medical word embeddings help the model to boost its performance for disease or syndrome, sign or symptom, and pharmacologic substance HNER tasks. We ranked the word embeddings by the performance as follows: (1) “Pyysalo Wiki + PM + PMC” achieved the highest result in 6/9 experiments, (2) “Chen PM + MIMIC III” achieved the highest result in 2/9 experiments, and (3) “Pyysalo PM + PMC” achieved the highest result in 1/9 experiments. Those three word embeddings are even more powerful than the rest of the embeddings together in the disease or syndrome, sign or symptom, and pharmacologic substance HNER with BiLSTM-CRF (word + char + POS) model.
The contribution of word embeddings to recognition of each named entity type is also different. “Chen PM + MIMIC-III” has more effect in recognition of disease or syndrome named entities than of the other named entities. “Pyysalo Wiki + PM + PMC” has more effects in the recognition of sign or symptom and pharmacologic substance named entities than of the other named entity.
We also examined the impact of fine-tuning embeddings in disease or syndrome, sign or symptom, and pharmacologic substance HNER by comparing the performance of BiLSTM-CRF (word + char + POS) model with that of an variant of it, in which “Pyysalo Wiki + PM + PMC” and “Chen PM + MIMIC-III” word embeddings are not fine-tuned during the model training as shown in
Table 9. The comparative results of two word embeddings with the model on the presented dataset demonstrate that fine-tuning embeddings has a certain effect on the performance of BiLSTM-CRF (word + char + POS) model. The F1 of BiLSTM-CRF with “Pyysalo Wiki + PM + PMC” is improved for disease or syndrome, sign or symptom, and pharmacologic substance HNER when the model uses fine-tuned embeddings, i.e., 0.99%, 1.45%, and 1.95%, respectively. The F1 of BiLSTM-CRF with “Chen PM + MIMIC III” is improved for disease or syndrome, sign or symptom, and pharmacologic substance HNER when the model uses fine-tuned embeddings, i.e., 0.39%, 1.16%, and 0.92%, respectively.
5. Conclusions
In this paper, we discuss advanced neural networks methods known as BiLSTM-CRF that are able to achieve the health-related NER task with word embedding, character embedding, and small feature engineering with POS tagging. The ontology or knowledge base is important for learning about the medical domain. Our goal is to predict and recognize medical terms in tweets that support public health systems. We annotated the collected dataset by using UMLS metathesaurus ontology to obtain knowledge about the specific domain. We considered three entity types: disease or syndrome, sign or symptom, and pharmacologic substance.
In the scope of HNER task, we presented a dataset collected from Twitter using the search term “healthcare” between 12 July 2018 and 12 July 2019, obtaining 676,251 tweets, 1,330,867 medical terms, and 19,727 unique medical terms. The presented dataset is larger than the previously presented dataset known as Micromed. The size of the dataset significantly improves the performance of the models. To produce the experimental dataset, we used the preprocessing techniques on the raw text data (tweets) such text cleaning, normalization, filtering, and removing non-medical terms and tokenization.
Inspired by this kind of work, we employed the BiLSTM-CRF model and compared with LSTM-CRF model with different combinations of features such as word embedding, character embedding, and POS tagging. Bidirectional models learn the input features in two ways: one from the beginning to end, and other from end to beginning, helping the learning of the feature more efficiently. We found that the BiLSTM-CRF (word + char + POS) model achieves the best result compared with other models on the HNER task when using “Pyysalo Wiki + PM + PMC” pre-trained word embeddings. The best model achieves a precision of 93.99%, recall of 73.31%, and F1-score of 81.77% for disease or syndrome HNER; a precision of 90.83%, recall of 81.98%, and F1-score of 87.52% for sign or symptom HNER; and a precision of 94.85%, recall of 73.47%, and F1-score of 84.51% for pharmacologic substance named entities. We also proved that fine-tuning is efficient when working on down-stream NLP tasks such as HNER.
As we found BiLSTM-CRF with “Pyysalo Wiki + PM + PMC” word embeddings, CNN-based character embedding and POS tagging is the best model for prediction of disease or syndrome, sign or symptom, and pharmacologic substance named entities.
In the future, we will extend the HNER task by adding different types of medical entities from UMLS entity types. We will apply transformer networks like BERT, ELMO, XLNET, etc. on the HNER tasks that currently dominate in most NLP tasks.