A Critical Assessment of the Congruency between Environmental DNA and Palaeoecology for the Biodiversity Monitoring and Palaeoenvironmental Reconstruction
Abstract
:1. Introduction
2. Methodology
2.1. Literature Search
2.2. PRISMA Analysis
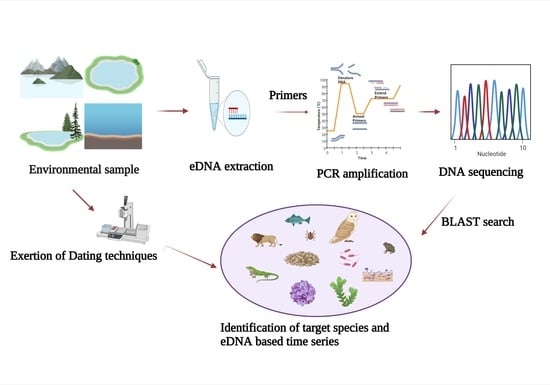
3. Sampling Design Optimization and Method Validation
3.1. Delineating the Sample Strategy
3.2. Sediment Coring
3.3. Inclusion of Appropriate Controls and Sample Preservation
4. Results and Discussion
4.1. Conceptual Background and Emergence of eDNA
4.2. Species Biomonitoring through Environmental DNA
| References | Habitat/Ecosystem | Representative Species | Use | Major Findings |
|---|---|---|---|---|
| Spear et al. (2021) [27] | Freshwater | Sander vitreus | Assessing population abundance | eDNA monitoring can appropriately dispense lakes to real world management categories for early warning for at-risk lakes in need of attention. |
| Afzali et al. (2021) [77] | Estuary | Demersal fish communities | Monitoring species biodiversity | eDNA metabarcoding out-competes traditional survey methods by enabling detection of rare and endangered taxa. |
| Boivin-Delisle et al. (2021) [28] | Freshwater | Sander vitreus | Species-specific biomonitoring | eDNA technique based on species-specific primers can provide insightful cognizance on fish biodiversity. |
| Polanco Fernández et al. (2021) [29] | Tropical marine coral reefs | Actinopterygii and Elasmobranchii | Species-specific biomonitoring | eDNA approach can provide an inclusive outline of fish composition in highly assorted coral reefs. |
| Capo et al. (2021) [17] | Lake sediments | Aquatic community | Biodiversity monitoring and palaeoenvironmental reconstructions | Despite a lack of clear and concise guidelines regarding sediment ancient DNA (SedaDNA), future SedaDNA research will provide more robust and result-oriented information about palaeoenvironments. |
| Thalinger et al. (2021) [18] | Riverine | Phoxinus phoxinus Salmo trutta Oncorhynchus mykiss Salvelinus fontinalis | Spatio-temporal shifts in ecosystem biodiversity | Seasonal discharge conditions prompt deep lateral and longitudinal changes in eDNA distribution. |
| Tsuji & Shibata (2021) [86] | Freshwater | Oryzias latipes Oryzias sakaizumii | Reproductive biology | Spawning events spike eDNA concentration, which offers the prospect to monitor and comprehend spawning timings with less effort than traditional methods. |
| Mejia et al. (2021) [19] | Desert springs | Plant and animal | Species recovery | eDNA is a promising supplemental tool to traditional approaches for biodiversity monitoring in desert springs. |
| Oka et al. (2021) [20] | Lagoon | Enneapterygius philippinus Spratelloides delicatulus Rhabdoblennius nitidus Enneapterygius similis | Biodiversity monitoring | For estimation of species diversity in tropical and subtropical areas, eDNA is a useful, rapid, and cost-effective method. |
| Székely et al. (2021) [21] | Arctic | Balaena mysticetus | Genetic diversity | Cetacean footprints are a promising cradle of genomic DNA. |
| Agerbo Rasmussen et al. (2021) [80] | Experimental vineyard | Fungi and arthropods | Species biomonitoring | eDNA offers a context for diversity assessment in vineyards to make more universal conclusions. |
| Shu, Ludwig, & Peng (2020) [73] | Freshwater | Freshwater fish Misgurnis anguillicaudates Cyprinus carpio Salvelinus fontinalis | Quantification | Despite its methodological obstacles, eDNA remains a promising and powerful contrivance for fish monitoring and conservation. |
| Zhang et al. (2020) [79] | Marine | Bacteria and marine mammals | Pelagic diversity | eDNA-based metabarcoding has the potential for successful multiple biodiversity surveillance, offering technical support and knowledge for future ecosystem protection and resource reservation. |
| Jeunen et al. (2019) [78] | Marine | Multi-specific | Species-specific biodiversity | The DNA extraction protocols when corrected and optimized provide clear illustration of eDNA monitoring in the marine environment. |
| Li et al. (2018) [85] | Freshwater | Invertebrates and human-induced contamination | Ecological monitoring | eDNA is not only applied for biodiversity monitoring but can be promising tool for understanding the impact of human-induced contamination in river ecosystems. |
| Ushio et al. (2018) [87] | Freshwater | Bird communities | Avian biodiversity patterns | eDNA metabarcoding method can serve as an essential alternative for taking a snapshot of bird diversity and potentially can be effective for ecosystem conservation and management. |
| Ramírez et al. (2018) [83] | Sediments | 16S rRNA extracellular genes | Biomonitoring | Extracellular 16S rRNA genes do not greatly influence the overall composition, abundance, and community richness. |
| Sansom & Sassoubre, (2017) [88] | Freshwater | Lampsilis siliquoidea | Quantification | eDNA approach holds tremendous potential for biomonitoring of species and can act as a complementary tool to protect the biodiversity. |
| Apothéloz-Perret-Gentil et al. (2017) [81] | Freshwater and streams | Epilithic samples | Benthic diatoms index | Taxonomy free molecular index can potentially extend its gauge and frequency to compliment current morphology-based methods for environmental biomonitoring. |
| Rees et al. (2017) [89] | Freshwater | Triturus cristatus | Species-specific identification | Environmental DNA has great proficiency and reproducibility in species-specific detection. |
| Klymus et al. (2017) [90] | Freshwater | Invasive species and native species | Biodiversity monitoring | The technique of eDNA can enhance identification and conservation efforts of native species and eradicating invasive species. |
| Deiner et al. (2016) [14] | Freshwater | Metazoan eukaryotes microinvertebrates | Biodiversity patterns | eDNA evaluates the biodiversity and ecological data over an entire landscape. |
| Guardiola et al. (2016) [33] | Marine | Deep-sea communities | Spatio-temporal biodiversity monitoring | eDNA can be a cornerstone for biomonitoring of deep-sea communities. |
| Valentini et al. (2016) [72] | Freshwater Marine | Amphibians bony fish | Aquatic biodiversity monitoring | For rare and secretive species, eDNA metabarcoding is the most proficient tool. Such an approach is crucial to address the fundamental and applied research question in ecology. |
| Thomsen et al. (2016) [22] | Sea water | Fish | Biodiversity monitoring | Application of eDNA for biodiversity assessment can be potentially beneficial not only for marine fish biomonitoring but also for science, society, and the global economy. |
| Davy et al. (2015) [91] | Freshwater | Sympatric turtles | Biomonitoring of threatened species | eDNA approach could provide a rapid and cost-effective alternative for the detection of freshwater turtles. |
| Willerslev et al. (2014) [94] | Arctic | Circumpolar plant diversity Nematode diversity | Arctic vegetation history by SedaDNA | eDNA in conjunction with dating methods can reflect information about the vegetation response to glacial climates. |
| Calvignac-Spencer et al. (2013) [23] | Forest | Mammalian diversity | Species biomonitoring | Caryion fly-derived DNA can be used to address the research questions pertaining to mammalian biodiversity. |
| Takahara et al. (2013) [92] | Ponds | Lepomis macrochirus | Distribution of invasive species | Distribution or presence of invasive species can be estimated more precisely based on eDNA as compared to traditional methods. |
| Taberlet et al. (2012) [93] | Soil Water | Multi-specific | Biodiversity assessment | Environmental DNA metabarcoding has massive potential to increase data acquisition in biodiversity exploration. |
| Dejean et al. (2012) [24] | Pond | American bullfrog | Species detection | eDNA method is valuable for species detection and surpasses survey methods in terms of sensitivity and sampling effort. |
| Darling & Mahon (2011) [42] | Freshwater | Invasive Asian carp | Biological invasion | eDNA technique is highly effective for the monitoring of aquatic invasive species. |
| Chariton et al. (2010) [25] | Estuarine sediments | Eukaryote ribosomal DNA | Ecological assessment | Next-generation pyrosequencing has the ability to identify and enumerate eukaryote species assemblages. |
| Hebsgaard et al. (2009) [26] | Permafrost | Dirt DNA | Archaeological context | Ancient DNA (aDNA) preserved in sediments can provide insights about the palaeoenvironmental conditions. |
| Ficetola et al. (2008) [57] | Freshwater | Frog (Rana catesbeiana) | Species-specific detection | Development of eDNA contrivance has opened new perspectives for biodiversity monitoring from environmental samples. |
4.3. Relation between eDNA and Palaeoenvironments
4.3.1. Lake Sediments
4.3.2. Permafrosts and Midden Material
5. Uncertainties Associated with eDNA Analysis and Potential Elucidations
6. The Way Forward
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barnosky, A.D.; Matzke, N.; Tomiya, S.; Wogan, G.O.; Swartz, B.; Quental, T.B.; Marshall, C.; McGuire, J.L.; Lindsey, E.L.; Maguire, K.C. Has the Earth’s sixth mass extinction already arrived? Nature 2011, 471, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Dirzo, R.; Young, H.S.; Galetti, M.; Ceballos, G.; Isaac, N.J.; Collen, B. Defaunation in the Anthropocene. Science 2014, 345, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Pimm, S.; Russell, G.; Gittleman, J.; Brooks, T. Futures. Biodivers. Sci. 1995, 269, 347–350. [Google Scholar]
- Ceballos, G.; Ehrlich, P.R.; Dirzo, R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc. Natl. Acad. Sci. USA 2017, 114, E6089–E6096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brondizio, E.S.; Settele, J.; Díaz, S.; Ngo, H.T. Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; IPBES Secretariat: Bonn, Germany, 2019. [Google Scholar]
- Maron, M.; Juffe-Bignoli, D.; Krueger, L.; Kiesecker, J.; Kümpel, N.F.; ten Kate, K.; Milner-Gulland, E.; Arlidge, W.N.; Booth, H.; Bull, J.W. Setting robust biodiversity goals. Conserv. Lett. 2021, 14, e12816. [Google Scholar] [CrossRef]
- Vié, J.-C.; Hilton-Taylor, C.; Stuart, S.N. Wildlife in a Changing World: An Analysis of the 2008 IUCN Red List of Threatened Species; IUCN: Fontainebleau, France, 2009. [Google Scholar]
- Abegão, J.L.R. Human Overpopulation Atlas. 2018. Available online: https://www.overpopulationatlas.com/ (accessed on 5 February 2020).
- Morita, K.; Sahashi, G.; Miya, M.; Kamada, S.; Kanbe, T.; Araki, H. Ongoing localized extinctions of stream-dwelling white-spotted charr populations in small dammed-off habitats of Hokkaido Island, Japan. Hydrobiologia 2019, 840, 207–213. [Google Scholar] [CrossRef]
- Niesenbaum, R.A. The integration of conservation, biodiversity, and sustainability. Sustainability 2019, 11, 4676. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Swihart, R.K. Absent or undetected? Effects of non-detection of species occurrence on wildlife–habitat models. Biol. Conserv. 2004, 116, 195–203. [Google Scholar] [CrossRef]
- MacKenzie, D.I.; Nichols, J.D.; Royle, J.A.; Pollock, K.H.; Bailey, L.; Hines, J.E. Occupancy Estimation and Modeling: Inferring Patterns and Dynamics of Species Occurrence; Elsevier: Burlington, MA, USA, 2017. [Google Scholar]
- Pawlowski, J.; Apothéloz-Perret-Gentil, L.; Altermatt, F. Environmental DNA: What’s behind the term? Clarifying the terminology and recommendations for its future use in biomonitoring. Mol. Ecol. 2020, 29, 4258–4264. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Bonin, A.; Coissac, E.; Zinger, L. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Cordier, T.; Alonso-Sáez, L.; Apothéloz-Perret-Gentil, L.; Aylagas, E.; Bohan, D.A.; Bouchez, A.; Chariton, A.; Creer, S.; Frühe, L.; Keck, F. Ecosystems monitoring powered by environmental genomics: A review of current strategies with an implementation roadmap. Mol. Ecol. 2021, 30, 2937–2958. [Google Scholar] [CrossRef]
- Capo, E.; Giguet-Covex, C.; Rouillard, A.; Nota, K.; Heintzman, P.D.; Vuillemin, A.; Ariztegui, D.; Arnaud, F.; Belle, S.; Bertilsson, S. Lake sedimentary DNA research on past terrestrial and aquatic biodiversity: Overview and recommendations. Quaternary 2021, 4, 6. [Google Scholar] [CrossRef]
- Thalinger, B.; Kirschner, D.; Pütz, Y.; Moritz, C.; Schwarzenberger, R.; Wanzenböck, J.; Traugott, M. Lateral and longitudinal fish environmental DNA distribution in dynamic riverine habitats. Environ. DNA 2021, 3, 305–318. [Google Scholar] [CrossRef]
- Palacios Mejia, M.; Curd, E.; Edalati, K.; Renshaw, M.A.; Dunn, R.; Potter, D.; Fraga, N.; Moore, J.; Saiz, J.; Wayne, R. The utility of environmental DNA from sediment and water samples for recovery of observed plant and animal species from four Mojave Desert springs. Environ. DNA 2021, 3, 214–230. [Google Scholar] [CrossRef]
- Oka, S.I.; Doi, H.; Miyamoto, K.; Hanahara, N.; Sado, T.; Miya, M. Environmental DNA metabarcoding for biodiversity monitoring of a highly diverse tropical fish community in a coral reef lagoon: Estimation of species richness and detection of habitat segregation. Environ. DNA 2021, 3, 55–69. [Google Scholar] [CrossRef]
- Székely, D.; Corfixen, N.L.; Mørch, L.L.; Knudsen, S.W.; McCarthy, M.L.; Teilmann, J.; Heide-Jørgensen, M.P.; Olsen, M.T. Environmental DNA captures the genetic diversity of bowhead whales (Balaena mysticetus) in West Greenland. Environ. DNA 2021, 3, 248–260. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Møller, P.R.; Sigsgaard, E.E.; Knudsen, S.W.; Jørgensen, O.A.; Willerslev, E. Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS ONE 2016, 11, e0165252. [Google Scholar] [CrossRef] [Green Version]
- Calvignac-Spencer, S.; Merkel, K.; Kutzner, N.; Kühl, H.; Boesch, C.; Kappeler, P.M.; Metzger, S.; Schubert, G.; Leendertz, F.H. Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Mol. Ecol. 2013, 22, 915–924. [Google Scholar] [CrossRef]
- Dejean, T.; Valentini, A.; Miquel, C.; Taberlet, P.; Bellemain, E.; Miaud, C. Improved detection of an alien invasive species through environmental DNA barcoding: The example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 2012, 49, 953–959. [Google Scholar] [CrossRef]
- Chariton, A.A.; Court, L.N.; Hartley, D.M.; Colloff, M.J.; Hardy, C.M. Ecological assessment of estuarine sediments by pyrosequencing eukaryotic ribosomal DNA. Front. Ecol. Environ. 2010, 8, 233–238. [Google Scholar] [CrossRef]
- Hebsgaard, M.B.; Gilbert, M.T.P.; Arneborg, J.; Heyn, P.; Allentoft, M.E.; Bunce, M.; Munch, K.; Schweger, C.; Willerslev, E. ‘The Farm Beneath the Sand’—An archaeological case study on ancient ‘dirt’DNA. Antiquity 2009, 83, 430–444. [Google Scholar] [CrossRef] [Green Version]
- Spear, M.J.; Embke, H.S.; Krysan, P.J.; Vander Zanden, M.J. Application of eDNA as a tool for assessing fish population abundance. Environ. DNA 2021, 3, 83–91. [Google Scholar] [CrossRef]
- Boivin-Delisle, D.; Laporte, M.; Burton, F.; Dion, R.; Normandeau, E.; Bernatchez, L. Using environmental DNA for biomonitoring of freshwater fish communities: Comparison with established gillnet surveys in a boreal hydroelectric impoundment. Environ. DNA 2021, 3, 105–120. [Google Scholar] [CrossRef]
- Polanco Fernández, A.; Marques, V.; Fopp, F.; Juhel, J.B.; Borrero-Pérez, G.H.; Cheutin, M.C.; Dejean, T.; González Corredor, J.D.; Acosta-Chaparro, A.; Hocdé, R. Comparing environmental DNA metabarcoding and underwater visual census to monitor tropical reef fishes. Environ. DNA 2021, 3, 142–156. [Google Scholar] [CrossRef]
- Ellegaard, M.; Clokie, M.R.; Czypionka, T.; Frisch, D.; Godhe, A.; Kremp, A.; Letarov, A.; McGenity, T.J.; Ribeiro, S.; John Anderson, N. Dead or alive: Sediment DNA archives as tools for tracking aquatic evolution and adaptation. Commun. Biol. 2020, 3, 169. [Google Scholar] [CrossRef] [Green Version]
- Prosser, J.I. Replicate or lie. Environ. Microbiol. 2010, 12, 1806–1810. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Guardiola, M.; Uriz, M.J.; Taberlet, P.; Coissac, E.; Wangensteen, O.S.; Turon, X. Deep-sea, deep-sequencing: Metabarcoding extracellular DNA from sediments of marine canyons. PLoS ONE 2015, 10, e0139633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Welch, D.M.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [Green Version]
- Leray, M.; Knowlton, N. DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proc. Natl. Acad. Sci. USA 2015, 112, 2076–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiiesalu, I.; Oepik, M.; Metsis, M.; Lilje, L.; Davison, J.; Vasar, M.; Moora, M.; Zobel, M.; Wilson, S.D.; Paertel, M. Plant species richness belowground: Higher richness and new patterns revealed by next-generation sequencing. Mol. Ecol. 2012, 21, 2004–2016. [Google Scholar] [CrossRef] [PubMed]
- Barberán, A.; McGuire, K.L.; Wolf, J.A.; Jones, F.A.; Wright, S.J.; Turner, B.L.; Essene, A.; Hubbell, S.P.; Faircloth, B.C.; Fierer, N. Relating belowground microbial composition to the taxonomic, phylogenetic, and functional trait distributions of trees in a tropical forest. Ecol. Lett. 2015, 18, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Bru, D.; Saby, N.P.; Čuhel, J.; Arrouays, D.; Šimek, M.; Hallin, S. Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ. Microbiol. 2009, 11, 3096–3104. [Google Scholar] [CrossRef]
- Albert, C.H.; Yoccoz, N.G.; Edwards, T.C., Jr.; Graham, C.H.; Zimmermann, N.E.; Thuiller, W. Sampling in ecology and evolution–bridging the gap between theory and practice. Ecography 2010, 33, 1028–1037. [Google Scholar] [CrossRef] [Green Version]
- Manel, S.; Albert, C.H.; Yoccoz, N.G. Sampling in landscape genomics. In Data Production and Analysis in Population Genomics; Springer: New York, NY, USA, 2012; pp. 3–12. [Google Scholar]
- Cao, Y.; Williams, D.D.; Larsen, D.P. Comparison of ecological communities: The problem of sample representativeness. Ecol. Monogr. 2002, 72, 41–56. [Google Scholar] [CrossRef]
- Darling, J.A.; Mahon, A.R. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Chadderton, W.L.; Mahon, A.R.; Renshaw, M.A.; Corush, J.; Budny, M.L.; Mysorekar, S.; Lodge, D.M. Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Can. J. Fish. Aquat. Sci. 2013, 70, 522–526. [Google Scholar] [CrossRef] [Green Version]
- Lafferty, K.D.; Benesh, K.C.; Mahon, A.R.; Jerde, C.L.; Lowe, C.G. Detecting southern California’s white sharks with environmental DNA. Front. Mar. Sci. 2018, 5, 355. [Google Scholar] [CrossRef]
- Jutzeler, M.; White, J.D.; Talling, P.J.; McCanta, M.; Morgan, S.; Le Friant, A.; Ishizuka, O. Coring disturbances in IODP piston cores with implications for offshore record of volcanic events and the Missoula megafloods. Geochem. Geophys. Geosyst. 2014, 15, 3572–3590. [Google Scholar] [CrossRef] [Green Version]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Willerslev, E.; Cappellini, E.; Boomsma, W.; Nielsen, R.; Hebsgaard, M.B.; Brand, T.B.; Hofreiter, M.; Bunce, M.; Poinar, H.N.; Dahl-Jensen, D. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science 2007, 317, 111–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willerslev, E.; Hansen, A.J.; Binladen, J.; Brand, T.B.; Gilbert, M.T.P.; Shapiro, B.; Bunce, M.; Wiuf, C.; Gilichinsky, D.A.; Cooper, A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 2003, 300, 791–795. [Google Scholar] [CrossRef] [Green Version]
- Kirkpatrick, J.B.; Walsh, E.A.; D’Hondt, S. Fossil DNA persistence and decay in marine sediment over hundred-thousand-year to million-year time scales. Geology 2016, 44, 615–618. [Google Scholar] [CrossRef] [Green Version]
- Bang-Andreasen, T.; Schostag, M.; Priemé, A.; Elberling, B.; Jacobsen, C.S. Potential microbial contamination during sampling of permafrost soil assessed by tracers. Sci. Rep. 2017, 7, 43338. [Google Scholar] [CrossRef] [PubMed]
- Willerslev, E.; Hansen, A.J.; Poinar, H.N. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol. Evol. 2004, 19, 141–147. [Google Scholar] [CrossRef]
- Wood, J.R.; Wilmshurst, J.M.; Wagstaff, S.J.; Worthy, T.H.; Rawlence, N.J.; Cooper, A. High-resolution coproecology: Using coprolites to reconstruct the habits and habitats of New Zealand’s extinct upland moa (Megalapteryx didinus). PLoS ONE 2012, 7, e40025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannoni, S.J.; Britschgi, T.B.; Moyer, C.L.; Field, K.G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 1990, 345, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Deagle, B.E.; Chiaradia, A.; McInnes, J.; Jarman, S.N. Pyrosequencing faecal DNA to determine diet of little penguins: Is what goes in what comes out? Conserv. Genet. 2010, 11, 2039–2048. [Google Scholar] [CrossRef]
- Pompanon, F.; Deagle, B.E.; Symondson, W.O.; Brown, D.S.; Jarman, S.N.; Taberlet, P. Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sønstebø, J.; Gielly, L.; Brysting, A.; Elven, R.; Edwards, M.; Haile, J.; Willerslev, E.; Coissac, E.; Rioux, D.; Sannier, J. Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Mol. Ecol. Resour. 2010, 10, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, C.S.; Pilliod, D.S.; Arkle, R.S.; Waits, L.P. Molecular detection of vertebrates in stream water: A demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE 2011, 6, e22746. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Koziol, A.; Stat, M.; Simpson, T.; Jarman, S.; DiBattista, J.D.; Harvey, E.S.; Marnane, M.; McDonald, J.; Bunce, M. Environmental DNA metabarcoding studies are critically affected by substrate selection. Mol. Ecol. Resour. 2019, 19, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, S.; Takahara, T.; Doi, H.; Shibata, N.; Yamanaka, H. The detection of aquatic macroorganisms using environmental DNA analysis—A review of methods for collection, extraction, and detection. Environ. DNA 2019, 1, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Tringe, S.G.; Von Mering, C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaalid, R.; Carlsen, T.; Kumar, S.; Halvorsen, R.; Ugland, K.I.; Fontana, G.; Kauserud, H. Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Mol. Ecol. 2012, 21, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 6213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kartzinel, T.R.; Chen, P.A.; Coverdale, T.C.; Erickson, D.L.; Kress, W.J.; Kuzmina, M.L.; Rubenstein, D.I.; Wang, W.; Pringle, R.M. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Natl. Acad. Sci. USA 2015, 112, 8019–8024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoccoz, N.G.; Bråthen, K.A.; Gielly, L.; Haile, J.; Edwards, M.E.; Goslar, T.; von Stedingk, H.; Brysting, A.; Coissac, E.; Pompanon, F.; et al. DNA from soil mirrors plant taxonomic and growth form diversity. Mol. Ecol. 2012, 21, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, D.S.; Colloff, M.J.; Rees, G.N.; Chariton, A.A.; Watson, G.O.; Court, L.N.; Hartley, D.M.; Morgan, M.J.; King, A.J.; Wilson, J.S. Impacts of inundation and drought on eukaryote biodiversity in semi-arid floodplain soils. Mol. Ecol. 2013, 22, 1746–1758. [Google Scholar] [CrossRef] [PubMed]
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 6237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Møller, P.R.; Rasmussen, M.; Willerslev, E. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 2012, 7, e41732. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for methods utilizing environmental DNA for detection of fish species. Genes 2020, 11, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epp, L.S.; Gussarova, G.; Boessenkool, S.; Olsen, J.; Haile, J.; Schrøder-Nielsen, A.; Ludikova, A.; Hassel, K.; Stenøien, H.K.; Funder, S. Lake sediment multi-taxon DNA from North Greenland records early post-glacial appearance of vascular plants and accurately tracks environmental changes. Quat. Sci. Rev. 2015, 117, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Giguet-Covex, C.; Pansu, J.; Arnaud, F.; Rey, P.-J.; Griggo, C.; Gielly, L.; Domaizon, I.; Coissac, E.; David, F.; Choler, P.; et al. Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nat. Commun. 2014, 5, 3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pansu, J.; Giguet-Covex, C.; Ficetola, G.F.; Gielly, L.; Boyer, F.; Zinger, L.; Arnaud, F.; Poulenard, J.; Taberlet, P.; Choler, P. Reconstructing long-term human impacts on plant communities: An ecological approach based on lake sediment DNA. Mol. Ecol. 2015, 24, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Afzali, S.F.; Bourdages, H.; Laporte, M.; Mérot, C.; Normandeau, E.; Audet, C.; Bernatchez, L. Comparing environmental metabarcoding and trawling survey of demersal fish communities in the Gulf of St. Lawrence, Canada. Environ. DNA 2021, 3, 22–42. [Google Scholar] [CrossRef]
- Jeunen, G.J.; Knapp, M.; Spencer, H.G.; Lamare, M.D.; Taylor, H.R.; Stat, M.; Bunce, M.; Gemmell, N.J. Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol. Ecol. Resour. 2019, 19, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, J.; Yao, M. A comprehensive and comparative evaluation of primers for metabarcoding eDNA from fish. Methods Ecol. Evol. 2020, 11, 1609–1625. [Google Scholar] [CrossRef]
- Agerbo Rasmussen, J.; Nielsen, M.; Mak, S.S.; Döring, J.; Klincke, F.; Gopalakrishnan, S.; Gilbert, M.T.P. eDNA-based biomonitoring at an experimental German vineyard to characterize how management regimes shape ecosystem diversity. Environ. DNA 2021, 3, 70–82. [Google Scholar] [CrossRef]
- Apothéloz-Perret-Gentil, L.; Cordonier, A.; Straub, F.; Iseli, J.; Esling, P.; Pawlowski, J. Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring. Mol. Ecol. Resour. 2017, 17, 1231–1242. [Google Scholar] [CrossRef]
- Capo, E.; Spong, G.; Koizumi, S.; Puts, I.; Olajos, F.; Königsson, H.; Karlsson, J.; Byström, P. Droplet digital PCR applied to environmental DNA, a promising method to estimate fish population abundance from humic-rich aquatic ecosystems. Environ. DNA 2021, 3, 343–352. [Google Scholar] [CrossRef]
- Ramírez, G.A.; Jørgensen, S.L.; Zhao, R.; D’Hondt, S. Minimal influence of extracellular DNA on molecular surveys of marine sedimentary communities. Front. Microbiol. 2018, 9, 2969. [Google Scholar] [CrossRef]
- Liang, R.; Li, Z.; Lau Vetter, M.C.; Vishnivetskaya, T.A.; Zanina, O.G.; Lloyd, K.G.; Pfiffner, S.M.; Rivkina, E.M.; Wang, W.; Wiggins, J.; et al. Genomic reconstruction of fossil and living microorganisms in ancient Siberian permafrost. Microbiome 2021, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Peng, Y.; Fang, W.; Altermatt, F.; Xie, Y.; Yang, J.; Zhang, X. Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers. Environ. Sci. Technol. 2018, 52, 11708–11719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, S.; Shibata, N. Identifying spawning events in fish by observing a spike in environmental DNA concentration after spawning. Environ. DNA 2021, 3, 190–199. [Google Scholar] [CrossRef]
- Ushio, M.; Murata, K.; Sado, T.; Nishiumi, I.; Takeshita, M.; Iwasaki, W.; Miya, M. Demonstration of the potential of environmental DNA as a tool for the detection of avian species. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansom, B.J.; Sassoubre, L.M. Environmental DNA (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environ. Sci. Technol. 2017, 51, 14244–14253. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.C.; Baker, C.A.; Gardner, D.S.; Maddison, B.C.; Gough, K.C. The detection of great crested newts year round via environmental DNA analysis. BMC Res. Notes 2017, 10, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klymus, K.E.; Marshall, N.T.; Stepien, C.A. Environmental DNA (eDNA) metabarcoding assays to detect invasive invertebrate species in the Great Lakes. PLoS ONE 2017, 12, e0177643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davy, C.M.; Kidd, A.G.; Wilson, C.C. Development and validation of environmental DNA (eDNA) markers for detection of freshwater turtles. PLoS ONE 2015, 10, e0130965. [Google Scholar] [CrossRef] [Green Version]
- Takahara, T.; Minamoto, T.; Doi, H. Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PLoS ONE 2013, 8, e56584. [Google Scholar]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef]
- Willerslev, E.; Davison, J.; Moora, M.; Zobel, M.; Coissac, E.; Edwards, M.E.; Lorenzen, E.D.; Vestergård, M.; Gussarova, G.; Haile, J. Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 2014, 506, 47–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawlence, N.J.; Lowe, D.J.; Wood, J.R.; Young, J.M.; Churchman, G.J.; Huang, Y.T.; Cooper, A. Using palaeoenvironmental DNA to reconstruct past environments: Progress and prospects. J. Quat. Sci. 2014, 29, 610–626. [Google Scholar] [CrossRef]
- Poinar, H.N.; Hofreiter, M.; Spaulding, W.G.; Martin, P.S.; Stankiewicz, B.A.; Bland, H.; Evershed, R.P.; Possnert, G.; Pääbo, S. Molecular coproscopy: Dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 1998, 281, 402–406. [Google Scholar] [CrossRef] [Green Version]
- Murchie, T.J. Ancient Environmental DNA as a Means of Understanding Ecological Restructuring during the Pleistocene-Holocene Transition in Yukon, Canada. Ph.D. Thesis, McMaster University, Hamilton, ON, Canada, 2021. [Google Scholar]
- Jones, G.; Jull, A.T.; Linick, T.; Donahue, D.J. Radiocarbon dating of deep-sea sediments: A comparison of accelerator mass spectrometer and beta-decay methods. Radiocarbon 1989, 31, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Stuiver, M. Tree ring, varve and carbon-14 chronologies. Nature 1970, 228, 454–455. [Google Scholar] [CrossRef] [PubMed]
- Jones, N. Carbon dating, the archaeological workhorse, is getting a major reboot. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.-L. The uranium-series methods of age determination. Annu. Rev. Earth Planet. Sci. 1976, 4, 347–379. [Google Scholar] [CrossRef]
- Chen, T.; Li, S.; Zhao, J.; Feng, Y. Uranium-thorium dating of coral mortality and community shift in a highly disturbed inshore reef (Weizhou Island, northern South China Sea). Sci. Total Environ. 2021, 752, 141866. [Google Scholar] [CrossRef]
- Berger, G.W. Dating Quaternary events by luminescence. In Dating Quaternary Sediments; Geological Society of America Boulder: Boulder, CO, USA, 1988; Volume 227, pp. 13–50. [Google Scholar]
- Hong, S.; Lee, M.K.; Seong, Y.B.; Owen, L.A.; Rhee, H.H.; Lee, J.I.; Yoo, K.-C. Holocene sea-level history and tectonic implications derived from luminescence dating of raised beaches in Terra Nova Bay, Antarctica. Geosci. J. 2021, 25, 283–298. [Google Scholar] [CrossRef]
- Aitken, M.J. Introduction to Optical Dating: The Dating of Quaternary Sediments by the Use of Photon-Stimulated Luminescence; Clarendon Press: Oxford, UK, 1998. [Google Scholar]
- Nian, X.; Zhang, W.; Wang, Z.; Sun, Q.; Chen, Z. Inter-comparison of optically stimulated luminescence (OSL) ages between different fractions of Holocene deposits from the Yangtze delta and its environmental implications. Mar. Geol. 2021, 432, 106401. [Google Scholar] [CrossRef]
- Nishiizumi, K.; Winterer, E.; Kohl, C.; Klein, J.; Middleton, R.; Lal, D.; Arnold, J. Cosmic ray production rates of 10Be and 26Al in quartz from glacially polished rocks. J. Geophys. Res. Solid Earth 1989, 94, 17907–17915. [Google Scholar] [CrossRef]
- Charreau, J.; Lave, J.; France-Lanord, C.; Puchol, N.; Blard, P.H.; Pik, R.; Gajurel, A.P.; ASTER Team. A 6 Ma record of palaeodenudation in the central Himalayas from in situ cosmogenic 10Be in the Surai section. Basin Res. 2021, 33, 1218–1239. [Google Scholar] [CrossRef]
- Cerling, T.E.; Craig, H. Geomorphology and in-situ cosmogenic isotopes. Annu. Rev. Earth Planet. Sci. 1994, 22, 273–317. [Google Scholar] [CrossRef]
- Yans, J.; Verhaert, M.; Gautheron, C.; Antoine, P.-O.; Moussi, B.; Dekoninck, A.; Decrée, S.; Chaftar, H.-R.; Hatira, N.; Dupuis, C. (U-Th)/He Dating of Supergene Iron (Oxyhydr-) Oxides of the Nefza-Sejnane District (Tunisia): New Insights into Mineralization and Mammalian Biostratigraphy. Minerals 2021, 11, 260. [Google Scholar] [CrossRef]
- Phillips, F.M.; Leavy, B.D.; Jannik, N.O.; Elmore, D.; Kubik, P.W. The accumulation of cosmogenic chlorine-36 in rocks: A method for surface exposure dating. Science 1986, 231, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Ram, R.; Purtschert, R.; Adar, E.M.; Bishof, M.; Jiang, W.; Lu, Z.-T.; Mueller, P.; Sy, A.; Vockenhuber, C.; Yechieli, Y.; et al. Controls on the 36Cl/Cl input ratio of paleo-groundwater in arid environments: New evidence from 81Kr/Kr data. Sci. Total Environ. 2021, 762, 144106. [Google Scholar] [CrossRef] [PubMed]
- Sarna-Wojcicki, A.; Lajoie, K.; Meyer, C.; Adam, D.; Rieck, H.; Morrison, R. Tephrochronologic correlation of upper Neogene sediments along the Pacific margin, conterminous United States. In Quaternary Nonglacial Geology; Geological Society of America, Inc.: Boulder, CO, USA, 1991; Volume 2, pp. 117–140. [Google Scholar]
- Westgate, J.A.; Gorton, M.P. Correlation techniques in tephra studies. In Tephra Studies; Springer: Dordrecht, The Netherlands, 1981; pp. 73–94. [Google Scholar]
- Smith, K.P.; Ólafsson, G.; Pálsdóttir, A.H. Ritual responses to catastrophic volcanism in Viking Age Iceland: Reconsidering Surtshellir Cave through Bayesian analyses of AMS dates, tephrochronology, and texts. J. Archaeol. Sci. 2021, 126, 105316. [Google Scholar] [CrossRef]
- Bada, J.L. The dating of fossil bones using the racemization of isoleucine. Earth Planet. Sci. Lett. 1972, 15, 223–231. [Google Scholar] [CrossRef]
- Bada, J.L.; Luyendyk, B.P.; Maynard, J.B. Marine sediments: Dating by the racemization of amino acids. Science 1970, 170, 730–732. [Google Scholar] [CrossRef]
- Wehmiller, J.F.; Belknap, D.F.; Boutin, B.S.; Mirecki, J.E.; Rahaim, S.D.; York, L.L. A review of the aminostratigraphy of Quaternary mollusks from United States Atlantic Coastal Plain sites. Geol. Soc. Am. Spec. Pap. 1988, 227, 69–110. [Google Scholar]
- Murray-Wallace, C.V.; Cann, J.H.; Yokoyama, Y.; Nicholas, W.A.; Lachlan, T.J.; Pan, T.-Y.; Dosseto, A.; Belperio, A.P.; Gostin, V.A. Late Pleistocene interstadial sea-levels (MIS 5a) in Gulf St Vincent, southern Australia, constrained by amino acid racemization dating of the benthic foraminifer Elphidium macelliforme. Quat. Sci. Rev. 2021, 259, 106899. [Google Scholar] [CrossRef]
- Cox, A.; Doell, R.R.; Dalrymple, G.B. Reversals of the earth’s magnetic field. Science 1964, 144, 1537–1543. [Google Scholar] [CrossRef]
- Rampino, M.R.; Shen, S.-Z. The end-Guadalupian (259.8 Ma) biodiversity crisis: The sixth major mass extinction? Hist. Biol. 2021, 33, 716–722. [Google Scholar] [CrossRef]
- Castelli, M.A.; Georges, A.; Cherryh, C.; Rosauer, D.F.; Sarre, S.D.; Contador-Kelsall, I.; Holleley, C.E. Evolving thermal thresholds explain the distribution of temperature sex reversal in an Australian dragon lizard. Divers. Distrib. 2021, 27, 427–438. [Google Scholar] [CrossRef]
- Creer, K. The dispersion of the geomagnetic field due to secular variation and its determination for remote times from paleomagnetic data. J. Geophys. Res. 1962, 67, 3461–3476. [Google Scholar] [CrossRef]
- Lund, S.P. A comparison of Holocene paleomagnetic secular variation records from North America. J. Geophys. Res. Solid Earth 1996, 101, 8007–8024. [Google Scholar] [CrossRef]
- Bieber, A.; St-Onge, G.; Feuillet, N.; Carlut, J.; Moreno, E.; Michel, E. Regional chronostratigraphy in the eastern Lesser Antilles quaternary fore-arc and accretionary wedge sediments: Relative paleointensity, oxygen isotopes and reversals. Quat. Geochronol. 2021, 65, 101179. [Google Scholar] [CrossRef]
- Fritts, H. Tree Rings and Climate; Academic Press: New York, NY, USA, 1976; p. 567. [Google Scholar]
- Jacoby, G.C.; Sheppard, P.R.; Sieh, K.E. Irregular recurrence of large earthquakes along the San Andreas fault: Evidence from trees. Science 1988, 241, 196–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, D.K.; Hoblitt, R.P. Tree-ring dating of pre-1980 volcanic flowage deposits at Mount St. Helens, Washington. Geol. Soc. Am. Bull. 1995, 107, 1077–1093. [Google Scholar] [CrossRef]
- Robinson, E.; Bocinsky, R.K.; Bird, D.; Freeman, J.; Kelly, R.L. Dendrochronological dates confirm a Late Prehistoric population decline in the American Southwest derived from radiocarbon dates. Philos. Trans. R. Soc. B 2021, 376, 20190718. [Google Scholar] [CrossRef]
- Suyama, Y.; Gunnarsson, U.; Parducci, L. Analysis of short DNA fragments from Holocene peatmoss samples. Holocene 2008, 18, 1003–1006. [Google Scholar] [CrossRef]
- Suyama, Y.; Kawamuro, K.; Kinoshita, I.; Yoshimura, K.; Tsumura, Y.; Takahara, H. DNA sequence from a fossil pollen of Abies spp. from Pleistocene peat. Genes Genet. Syst. 1996, 71, 145–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, K.; Zhang, J.; He, K.; Wang, C.; Lu, H. Impacts of the Wetland Environment on Demographic Development during the Neolithic in the Lower Yangtze Region—Based on Peat and Archaeological Dates. Front. Earth Sci. 2021, 9, 59. [Google Scholar] [CrossRef]
- Arnold, L.J.; Roberts, R.G.; MacPhee, R.D.; Haile, J.S.; Brock, F.; Möller, P.; Froese, D.G.; Tikhonov, A.N.; Chivas, A.R.; Gilbert, M.T.P. Paper II–dirt, dates and DNA: OSL and radiocarbon chronologies of perennially frozen sediments in Siberia, and their implications for sedimentary ancient DNA studies. Boreas 2011, 40, 417–445. [Google Scholar] [CrossRef]
- Johnson, S.S.; Hebsgaard, M.B.; Christensen, T.R.; Mastepanov, M.; Nielsen, R.; Munch, K.; Brand, T.; Gilbert, M.T.P.; Zuber, M.T.; Bunce, M. Ancient bacteria show evidence of DNA repair. Proc. Natl. Acad. Sci. USA 2007, 104, 14401–14405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, T.; Reeves, R.; Gilichinsky, D.; Friedmann, E. Characterization of viable bacteria from Siberian permafrost by 16S rDNA sequencing. Microb. Ecol. 1997, 33, 169–179. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Petrova, M.A.; Urbance, J.; Ponder, M.; Moyer, C.L.; Gilichinsky, D.A.; Tiedje, J.M. Bacterial community in ancient Siberian permafrost as characterized by culture and culture-independent methods. Astrobiology 2006, 6, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Massicotte, P.; Amon, R.M.; Antoine, D.; Archambault, P.; Balzano, S.; Bélanger, S.; Benner, R.; Boeuf, D.; Bricaud, A.; Bruyant, F. The MALINA oceanographic expedition: How do changes in ice cover, permafrost and UV radiation impact biodiversity and biogeochemical fluxes in the Arctic Ocean? Earth Syst. Sci. Data 2021, 13, 1561–1592. [Google Scholar] [CrossRef]
- Murchie, T.J.; Kuch, M.; Duggan, A.T.; Ledger, M.L.; Roche, K.; Klunk, J.; Karpinski, E.; Hackenberger, D.; Sadoway, T.; MacPhee, R. Optimizing extraction and targeted capture of ancient environmental DNA for reconstructing past environments using the PalaeoChip Arctic-1.0 bait-set. Quat. Res. 2021, 99, 305–328. [Google Scholar] [CrossRef]
- Varotto, C.; Pindo, M.; Bertoni, E.; Casarotto, C.; Camin, F.; Girardi, M.; Maggi, V.; Cristofori, A. A pilot study of eDNA metabarcoding to estimate plant biodiversity by an alpine glacier core (Adamello Glacier, North Italy). Sci. Rep. 2021, 11, 1208. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; Rogers, S.O.; Catranis, C.M.; Starmer, W.T. Detection and characterization of ancient fungi entrapped in glacial ice. Mycologia 2000, 92, 286–295. [Google Scholar] [CrossRef]
- Tian, Y.; Spicer, R.A.; Huang, J.; Zhou, Z.; Su, T.; Widdowson, M.; Jia, L.; Li, S.; Wu, W.; Xue, L. New early oligocene zircon U-Pb dates for the ‘Miocene’ Wenshan Basin, Yunnan, China: Biodiversity and paleoenvironment. Earth Planet. Sci. Lett. 2021, 565, 116929. [Google Scholar] [CrossRef]
- Ingels, J.; Aronson, R.B.; Smith, C.R.; Baco, A.; Bik, H.M.; Blake, J.A.; Brandt, A.; Cape, M.; Demaster, D.; Dolan, E. Antarctic ecosystem responses following ice-shelf collapse and iceberg calving: Science review and future research. Wiley Interdiscip. Rev. Clim. Chang. 2021, 12, e682. [Google Scholar] [CrossRef]
- Bennett, K.D.; Parducci, L. DNA from pollen: Principles and potential. Holocene 2006, 16, 1031–1034. [Google Scholar] [CrossRef] [Green Version]
- Gugerli, F.; Alvarez, N.; Tinner, W. A deep dig—Hindsight on Holocene vegetation composition from ancient environmental DNA. Mol. Ecol. 2013, 22, 3433–3436. [Google Scholar] [CrossRef] [PubMed]
- Parducci, L.; Suyama, Y.; Lascoux, M.; Bennett, K.D. Ancient DNA from pollen: A genetic record of population history in Scots pine. Mol. Ecol. 2005, 14, 2873–2882. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.W.; Ginolhac, A.; Orlando, L.; Olsen, J.; Andersen, K.; Holm, J.; Funder, S.; Willerslev, E.; Kjær, K.H. A comparative study of ancient environmental DNA to pollen and macrofossils from lake sediments reveals taxonomic overlap and additional plant taxa. Quat. Sci. Rev. 2013, 75, 161–168. [Google Scholar] [CrossRef]
- Xu, Z.-H.; Jiang, X.-D.; Wang, G.-Z.; He, J.-F.; Cai, M.-H.; Wu, L.-S.; Jiang, J.-L.; Chen, X.-L. DNA extraction, amplification and analysis of the 28S rRNA portion in sediment-buried copepod DNA in the Great Wall Bay and Xihu Lake, Antarctica. J. Plankton Res. 2011, 33, 917–925. [Google Scholar] [CrossRef] [Green Version]
- Rudaya, N.; Nazarova, L.; Frolova, L.; Palagushkina, O.; Soenov, V.; Cao, X.; Syrykh, L.; Grekov, I.; Otgonbayar, D.; Bayarkhuu, B. The link between climate change and biodiversity of lacustrine inhabitants and terrestrial plant communities of the Uvs Nuur Basin (Mongolia) during the last three millennia. Holocene 2021, 31, 09596836211019093. [Google Scholar] [CrossRef]
- Zhang, H.; Huo, S.; Yeager, K.M.; Wu, F. Sedimentary DNA record of eukaryotic algal and cyanobacterial communities in a shallow Lake driven by human activities and climate change. Sci. Total Environ. 2021, 753, 141985. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Elias, S.; Gilbert, M.T.P.; Haile, J.; Munch, K.; Kuzmina, S.; Froese, D.G.; Sher, A.; Holdaway, R.N.; Willerslev, E. Non-destructive sampling of ancient insect DNA. PLoS ONE 2009, 4, e5048. [Google Scholar] [CrossRef]
- Lazagabaster, I.A.; Ullman, M.; Porat, R.; Halevi, R.; Porat, N.; Davidovich, U.; Marom, N. Changes in the large carnivore community structure of the Judean Desert in connection to Holocene human settlement dynamics. Sci. Rep. 2021, 11, 3548. [Google Scholar] [CrossRef] [PubMed]
- David, B.; Arnold, L.J.; Delannoy, J.-J.; Fresløv, J.; Urwin, C.; Petchey, F.; McDowell, M.C.; Mullett, R.; Land, G.; Mialanes, J. Late survival of megafauna refuted for Cloggs Cave, SE Australia: Implications for the Australian Late Pleistocene megafauna extinction debate. Quat. Sci. Rev. 2021, 253, 106781. [Google Scholar] [CrossRef]
- Caldeira, A.T.; Schiavon, N.; Mauran, G.; Salvador, C.; Rosado, T.; Mirão, J.; Candeias, A. On the Biodiversity and Biodeteriogenic Activity of Microbial Communities Present in the Hypogenic Environment of the Escoural Cave, Alentejo, Portugal. Coatings 2021, 11, 209. [Google Scholar] [CrossRef]
- Paffetti, D.; Vettori, C.; Caramelli, D.; Vernesi, C.; Lari, M.; Paganelli, A.; Paule, L.; Giannini, R. Unexpected presence of Fagus orientalis complex in Italy as inferred from 45,000-year-old DNA pollen samples from Venice lagoon. BMC Evol. Biol. 2007, 7, S6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albano, P.G.; Steger, J.; Bošnjak, M.; Dunne, B.; Guifarro, Z.; Turapova, E.; Hua, Q.; Kaufman, D.S.; Rilov, G.; Zuschin, M. Native biodiversity collapse in the eastern Mediterranean. Proc. R. Soc. B 2021, 288, 20202469. [Google Scholar] [CrossRef] [PubMed]
- Alves, E.Q.; Macario, K.D.; Spotorno, P.; Oliveira, F.M.; Muniz, M.C.; Fallon, S.; Souza, R.; Salvador, A.; Eschner, A.; Ramsey, C.B. Nineteenth-century expeditions and the radiocarbon marine reservoir effect on the Brazilian coast. Geochim. Cosmochim. Acta 2021, 297, 276–287. [Google Scholar] [CrossRef]
- Quarta, G.; Maruccio, L.; D’Elia, M.; Calcagnile, L. Radiocarbon Dating of Marine Samples: Methodological Aspects, Applications and Case Studies. Water 2021, 13, 986. [Google Scholar] [CrossRef]
- Vieira, C.; Antoine De Ramon, N.Y.; Rasoamanendrika, F.A.; D’Hondt, S.; Tran, L.-A.T.; Van den Spiegel, D.; Kawai, H.; De Clerck, O. Marine macroalgal biodiversity of northern Madagascar: Morpho-genetic systematics and implications of anthropic impacts for conservation. Biodivers. Conserv. 2021, 30, 1501–1546. [Google Scholar] [CrossRef]
- Gould, B.A.; León, B.; Buffen, A.M.; Thompson, L.G. Evidence of a high-Andean, mid-Holocene plant community: An ancient DNA analysis of glacially preserved remains. Am. J. Bot. 2010, 97, 1579–1584. [Google Scholar] [CrossRef] [Green Version]
- Schlütz, F.; Lehmkuhl, F. Holocene climatic change and the nomadic Anthropocene in Eastern Tibet: Palynological and geomorphological results from the Nianbaoyeze Mountains. Quat. Sci. Rev. 2009, 28, 1449–1471. [Google Scholar] [CrossRef]
- Oliva, M.; Ruiz-Fernández, J. Late Quaternary environmental dynamics in Lenin Peak area (Pamir Mountains, Kyrgyzstan). Sci. Total Environ. 2018, 645, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Falardeau, J.; de Vernal, A.; Seidenkrantz, M.-S.; Fritz, M. Benthic foraminifer and ostracod assemblages in the Beaufort Sea continental shelf over the last millennia: Evidence of unprecedented changes in the last two centuries. In Proceedings of the PALEOARC 2021—2nd International Conference on ‘Processes and Palaeo-Environmental Changes in the Arctic from Past to Present’, Online, 25–28 May 2021. [Google Scholar]
- Jørgensen, T.; Kjaer, K.H.; Haile, J.; Rasmussen, M.; Boessenkool, S.; Andersen, K.; Coissac, E.; Taberlet, P.; Brochmann, C.; Orlando, L. Islands in the ice: Detecting past vegetation on Greenlandic nunataks using historical records and sedimentary ancient DNA Meta-barcoding. Mol. Ecol. 2012, 21, 1980–1988. [Google Scholar] [CrossRef] [PubMed]
- Wilmshurst, J.M.; Moar, N.T.; Wood, J.R.; Bellingham, P.J.; Findlater, A.M.; Robinson, J.J.; Stone, C. Use of pollen and ancient DNA as conservation baselines for offshore islands in New Zealand. Conserv. Biol. 2014, 28, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Delgado-Baquerizo, M.; Guo, X.; Wang, D.; Zhu, Y.-G.; Chu, H. Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J. 2021, 15, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Guerra, C.A.; Bardgett, R.D.; Caon, L.; Crowther, T.W.; Delgado-Baquerizo, M.; Montanarella, L.; Navarro, L.M.; Orgiazzi, A.; Singh, B.K.; Tedersoo, L. Tracking, targeting, and conserving soil biodiversity. Science 2021, 371, 239–241. [Google Scholar] [CrossRef]
- Jou, R.M.; Macario, K.D.; Pessenda, L.C.; Pereira, M.G.; Lorente, F.L.; Pedrosa, R.; da Silva Neto, E.C.; Fallon, S.; Muniz, M.C.; Cardoso, R.P. The use of carbon isotopes (13C, 14C) in different soil types and vegetation coverage in a montane atlantic forest region, Southeast Brazil. Quat. Geochronol. 2021, 61, 101133. [Google Scholar] [CrossRef]
- Kuch, M.; Poinar, H. Extraction of DNA from paleofeces. In Ancient DNA; Springer: New York, NY, USA, 2012; pp. 37–42. [Google Scholar]
- Merklinger, F.F.; Böhnert, T.; Arakaki, M.; Weigend, M.; Quandt, D.; Luebert, F. Quaternary diversification of a columnar cactus in the driest place on earth. Am. J. Bot. 2021, 108, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Oswald, J.A.; Terrill, R.S.; Stucky, B.J.; LeFebvre, M.J.; Steadman, D.W.; Guralnick, R.P.; Allen, J.M. Ancient DNA from the extinct Haitian cave-rail (Nesotrochis steganinos) suggests a biogeographic connection between the Caribbean and Old World. Biol. Lett. 2021, 17, 20200760. [Google Scholar] [CrossRef]
- Rabinow, S.; Giovas, C. A systematic review of agouti (Dasyproctidae: Dasyprocta) records from the pre-1492 Lesser Antilles: New perspectives on an introduced commensal. Int. J. Osteoarchaeol. 2021, 31, 758–769. [Google Scholar] [CrossRef]
- Witt, K.E.; Yarlagadda, K.; Allen, J.M.; Bader, A.C.; Simon, M.L.; Kuehn, S.R.; Swanson, K.S.; Cross, T.-W.L.; Hedman, K.M.; Ambrose, S.H.; et al. Integrative analysis of DNA, macroscopic remains and stable isotopes of dog coprolites to reconstruct community diet. Sci. Rep. 2021, 11, 3113. [Google Scholar] [CrossRef] [PubMed]
- Leung, T.L. Parasites of Fossil Vertebrates: What We Know and What Can We Expect from the Fossil Record? In The Evolution and Fossil Record of Parasitism: Identification and Macroevolution of Parasites; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Rummy, P.; Halaclar, K.; Chen, H. The first record of exceptionally-preserved spiral coprolites from the Tsagan-Tsab formation (lower cretaceous), Tatal, western Mongolia. Sci. Rep. 2021, 11, 7891. [Google Scholar] [CrossRef] [PubMed]
- Van Geel, B.; Aptroot, A.; Baittinger, C.; Birks, H.H.; Bull, I.D.; Cross, H.B.; Evershed, R.P.; Gravendeel, B.; Kompanje, E.J.; Kuperus, P. The ecological implications of a Yakutian mammoth’s last meal. Quat. Res. 2008, 69, 361–376. [Google Scholar] [CrossRef] [Green Version]
- Wood, J.R.; Rawlence, N.J.; Rogers, G.M.; Austin, J.J.; Worthy, T.H.; Cooper, A. Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes). Quat. Sci. Rev. 2008, 27, 2593–2602. [Google Scholar] [CrossRef]
- Wood, J.R.; Wilmshurst, J.M.; Richardson, S.J.; Rawlence, N.J.; Wagstaff, S.J.; Worthy, T.H.; Cooper, A. Resolving lost herbivore community structure using coprolites of four sympatric moa species (Aves: Dinornithiformes). Proc. Natl. Acad. Sci. USA 2013, 110, 16910–16915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parducci, L.; Matetovici, I.; Fontana, S.L.; Bennett, K.D.; Suyama, Y.; Haile, J.; Kjær, K.H.; Larsen, N.K.; Drouzas, A.D.; Willerslev, E. Molecular-and pollen-based vegetation analysis in lake sediments from central Scandinavia. Mol. Ecol. 2013, 22, 3511–3524. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Edwards, M.E. The maturing relationship between Quaternary paleoecology and ancient sedimentary DNA. Quat. Res. 2020, 96, 39–47. [Google Scholar] [CrossRef]
- Sawyer, S.; Krause, J.; Guschanski, K.; Savolainen, V.; Pääbo, S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE 2012, 7, e34131. [Google Scholar]
- Richards, G. Human Evolution: An Introduction for the Behavioural Sciences; Routledge: Oxford, UK, 2019. [Google Scholar]
- Schnell, I.B.; Thomsen, P.F.; Wilkinson, N.; Rasmussen, M.; Jensen, L.R.; Willerslev, E.; Bertelsen, M.F.; Gilbert, M.T.P. Screening mammal biodiversity using DNA from leeches. Curr. Biol. 2012, 22, R262–R263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seersholm, F.V.; Cole, T.L.; Grealy, A.; Rawlence, N.J.; Greig, K.; Knapp, M.; Stat, M.; Hansen, A.J.; Easton, L.J.; Shepherd, L. Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey. Proc. Natl. Acad. Sci. USA 2018, 115, 7771–7776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, C.S.; Sepulveda, A.; Ray, A.; Baumgardt, J.; Waits, L.P. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw. Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef] [Green Version]
- Clare, E.L.; Chain, F.J.; Littlefair, J.E.; Cristescu, M.E. The effects of parameter choice on defining molecular operational taxonomic units and resulting ecological analyses of metabarcoding data. Genome 2016, 59, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Creer, S.; Deiner, K.; Frey, S.; Porazinska, D.; Taberlet, P.; Thomas, W.K.; Potter, C.; Bik, H.M. The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol. Evol. 2016, 7, 1008–1018. [Google Scholar] [CrossRef]
- Piggott, M.P. Evaluating the effects of laboratory protocols on eDNA detection probability for an endangered freshwater fish. Ecol. Evol. 2016, 6, 2739–2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, R.; Zhan, A.; Brown, E.A.; Chain, F.J.; Cristescu, M.E.; Gras, R.; MacIsaac, H.J. Optimization and performance testing of a sequence processing pipeline applied to detection of nonindigenous species. Evol. Appl. 2018, 11, 891–905. [Google Scholar] [CrossRef] [Green Version]
- Coissac, E.; Riaz, T.; Puillandre, N. Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 2012, 21, 1834–1847. [Google Scholar] [CrossRef]
- Roussel, J.M.; Paillisson, J.M.; Treguier, A.; Petit, E. The downside of eDNA as a survey tool in water bodies. J. Appl. Ecol. 2015, 52, 823–826. [Google Scholar] [CrossRef]
- Cristescu, M.E. From barcoding single individuals to metabarcoding biological communities: Towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 2014, 29, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Kraaijeveld, K.; De Weger, L.A.; Ventayol García, M.; Buermans, H.; Frank, J.; Hiemstra, P.S.; Den Dunnen, J.T. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Mol. Ecol. Resour. 2015, 15, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Cordier, T.; Esling, P.; Lejzerowicz, F.; Visco, J.; Ouadahi, A.; Martins, C.; Cedhagen, T.; Pawlowski, J. Predicting the ecological quality status of marine environments from eDNA metabarcoding data using supervised machine learning. Environ. Sci. Technol. 2017, 51, 9118–9126. [Google Scholar] [CrossRef] [PubMed]
- Tournayre, O.; Leuchtmann, M.; Galan, M.; Trillat, M.; Piry, S.; Pinaud, D.; Filippi-Codaccioni, O.; Pontier, D.; Charbonnel, N. eDNA metabarcoding reveals a core and secondary diets of the greater horseshoe bat with strong spatio-temporal plasticity. Environ. DNA 2021, 3, 277–296. [Google Scholar] [CrossRef]





| Search Word | Search Field | Number of Hits in Major Data Bases | Last Updated | |
|---|---|---|---|---|
| PubMed | Scopus | |||
| “eDNA” | Article, title, keywords | 1270 | 1640 | 31 July 2021 |
| “eDNA and aquatic” | Article, title, keywords | 290 | 169 | 31 July 2021 |
| “eDNA and freshwater” | Article, title, keywords | 141 | 157 | 31 July 2021 |
| “eDNA and sediments” | Article, title, keywords | 51 | 71 | 31 July 2021 |
| “eDNA and diversity” | Article, title, keywords | 125 | 163 | 31 July 2021 |
| “eDNA and palaeoenvironmental reconstructions” | Article, title, keywords | 39 | 59 | 31 July 2021 |
| Method | Range | Materials | References |
|---|---|---|---|
| Radioisotopic | |||
| 14C | 35 ka | wood, shell | [98,99,100] |
| U/Th | 10–350 ka | Carbonate (corals, speleothems) | [101,102] |
| Thermoluminescence (TL) | 30–300 ka | quartz silt | [103,104] |
| Optically Stimulated Luminescence | 0–300 ka | quartz silt | [105,106] |
| Cosmogenic | |||
| In situ 10Be, 26Al | 3–4 Ma | Quartz | [107,108] |
| He, Ne | Unlimited | [109,110] | |
| 36Cl | 0–4 Ma | Olivine, quartz | [111,112] |
| Chemical | |||
| Tephrochronology | 0–several Ma | Volcanic ash | [113,114,115] |
| Amino acid racemization | 0–300 ka; range temperature reliant | Carbonate shell | [116,117,118,119] |
| Paleomagnetic | |||
| Identification of reversals | >700 ka | Fine sediments, volcanic flows | [120,121,122] |
| Secular versions | 0–700 ka | Fine sediments | [123,124,125] |
| Biological | |||
| Dendrochronology | 10 ka, subject to indigenous master chronology | Wood | [126,127,128,129] |
| Material | Target Taxa | Age Range | References |
|---|---|---|---|
| Peat | Plantae | 155 ka | [130,131,132] |
| Permafrost | Bacteria, fungi, bryophyta, plantae, insecta, mammalia, aves | 2–<600 ka | [133,134,135,136,137,138,139] |
| Ice | Fungi, protista, plantae, insect | 0.3–<800 ka | [47,139,140,141,142] |
| Lacustrine | Diatoms, plantae, crustacea, copepod | 13 cal ka–modern | [143,144,145,146,147,148,149] |
| Cave deposits | Plantae, insecta, mammalia, aves | 10.8–0.6 14C ka | [150,151,152,153] |
| Marine | Foraminifera, radiolarian, plantae | ≤45 ka | [154,155,156,157,158] |
| Glacial (fluviogravel and moraine) | Plantae | 4.5–5.2 cal ka | [159,160,161,162] |
| Soil | Plantae, mammalia, Aves | 5.5 cal ka–modern | [163,164,165,166,167] |
| Rodent, midden | Plantae, vertebrata | 10.1 14C ka | [168,169,170,171] |
| Coprolites | Plantae, parasites, mammalia, aves | 32–06 14C ka | [164,172,173,174] |
| Problem/Limitation | Elucidation of Problem | Methods Affected | Possible Solutions | References |
|---|---|---|---|---|
| Wrong method | Detection possibility of eDNA and estimation of biodiversity is affected by field, laboratory, and bioinformatics protocols, thereby making it obligatory to select an optimal protocol as imperfectly designed methodology impacts the results. | Microarrays, PCR, qPCR, and Metabarcoding | Execution of the relative field findings and the laboratory procedures. Adoption of validated and justified means through exploration of mock communities | [186,187,188,189] |
| False positives | Improper handling of samples, lacking adequate specificity of primers and probes, errors in data analysis, and mutations that accumulate post-mortem produce false positives. | Microarrays PCR qPCR Metabarcoding | Include positive and negative controls for appropriate optimization of protocols, expending numerous markers or primers, choose suitable factors for bioinformatics scrutiny of sequences. | [35,42,190,191] |
| False negatives | False negatives arise owing to prompt degradation or limited eDNA amount in samples. Primer bias can also result in false negatives. | Microarrays PCR qPCR Metabarcoding | To confirm the sampling size, the accumulation curve of species can be generated to attain an asymptote; furthermore, multiple PCRs on each extract can be conducted, appraisal of results in contrast to customary community composition assessments. | [42,187,190,191,192,193] |
| Partial ecological evidence | Owing to deficiency of information about the sex and size of the individuals distinguished by eDNA. | Microarrays PCR qPCR Metabarcoding | Life stage and sex certain markers can be used to overcome limitations of such nature. | [72] |
| Limited taxonomic resolution | Uncategorized diversity and its poor linkage to their ecology. | Barcoding Metabarcoding | When few target species are investigated, utilize local reference libraries. Employ global reference sequence | [35,193,194] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, S.; Khurshid, Z.; Sabreena; Bali, B.S.; Ganai, B.A.; Sayyed, R.Z.; Poczai, P.; Zaman, M. A Critical Assessment of the Congruency between Environmental DNA and Palaeoecology for the Biodiversity Monitoring and Palaeoenvironmental Reconstruction. Int. J. Environ. Res. Public Health 2022, 19, 9445. https://doi.org/10.3390/ijerph19159445
Hassan S, Khurshid Z, Sabreena, Bali BS, Ganai BA, Sayyed RZ, Poczai P, Zaman M. A Critical Assessment of the Congruency between Environmental DNA and Palaeoecology for the Biodiversity Monitoring and Palaeoenvironmental Reconstruction. International Journal of Environmental Research and Public Health. 2022; 19(15):9445. https://doi.org/10.3390/ijerph19159445
Chicago/Turabian StyleHassan, Shahnawaz, Zulaykha Khurshid, Sabreena, Bikram Singh Bali, Bashir Ah Ganai, R. Z. Sayyed, Peter Poczai, and Muzafar Zaman. 2022. "A Critical Assessment of the Congruency between Environmental DNA and Palaeoecology for the Biodiversity Monitoring and Palaeoenvironmental Reconstruction" International Journal of Environmental Research and Public Health 19, no. 15: 9445. https://doi.org/10.3390/ijerph19159445
APA StyleHassan, S., Khurshid, Z., Sabreena, Bali, B. S., Ganai, B. A., Sayyed, R. Z., Poczai, P., & Zaman, M. (2022). A Critical Assessment of the Congruency between Environmental DNA and Palaeoecology for the Biodiversity Monitoring and Palaeoenvironmental Reconstruction. International Journal of Environmental Research and Public Health, 19(15), 9445. https://doi.org/10.3390/ijerph19159445