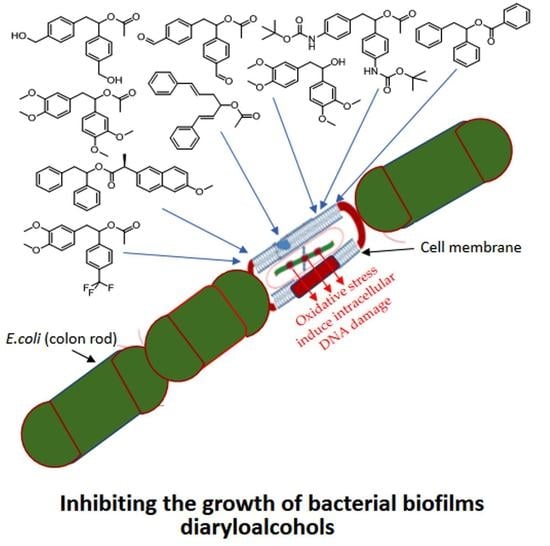
1,2-Diarylethanols—A New Class of Compounds That Are Toxic to E. coli K12, R2–R4 Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Media
2.2. Experimental Chemistry
- General Scheme of Reaction Synthesis for Compounds 1–18
- General Procedure for the Synthesis of 1,2-Diarylethyl Esters 4
- General Procedure for the Hydrolysis of 1,2-Diarylethyl Ester 4 to Alcohols 7
2.2.1. Product 4a 1,2-Diphenylethyl Acetate
2.2.2. Product 4b 1,2-di-p-Tolylethyl Acetate
2.2.3. Product 4c 1,2-bis(4-(Trifluoromethyl)phenyl)ethyl Acetate
2.2.4. Product 4d 1,2-bis(4-Chlorophenyl)ethyl Acetate
2.2.5. Product 4e 1,2-bis(4-Bromophenyl)ethyl Acetate
2.2.6. Product 4f 1,2-bis(4-Iodophenyl)ethyl Acetate
2.2.7. Product 4h 1,2-bis(4-Formylphenyl)ethyl Acetate
2.2.8. Product 4i 1,2-bis(4-(Hydroxymethyl)phenyl)ethyl Acetate
2.2.9. Product 4j 1,2-bis(4-Methoxyphenyl)ethyl Acetate
2.2.10. Product 4k 1,2-bis(3,4-Dimethoxyphenyl)ethyl Acetate
2.2.11. Product 4l 1,2-bis(4-((tert-Butoxycarbonyl)amino)phenyl)ethyl Acetate
2.2.12. Product 4la (1E,5E)-1,6-Diphenylhexa-1,5-dien-3-yl acetate
2.2.13. Product 4ck (3,4-Dimethoxyphenyl)-1-(4-(Trifluoromethyl)phenyl)ethyl Acetate
2.2.14. Product 4q 1,2-Diphenylethyl Benzoate
2.2.15. Product 4t 1,2-Diphenylethyl 2-Chloroacetate
2.2.16. Product 4u (2S)-1,2-Diphenylethyl 2-(6-Methoxynaphthalen-2-yl)propanoate
2.2.17. Product 7a 1,2-Diphenylethanol
2.2.18. Product 7k 1,2-bis(3,4-Dimethoxyphenyl)ethanol
2.3. Application of MIC and MBC Tests
2.4. Isolation Plasmids DNA from Bacterial K12, R2–R4 Strains
2.4.1. Interaction of the Plasmid DNA from K12 and R4 Strains with 1,2-Diaryloethanols
2.4.2. Interaction of the Plasmid DNA from K12 and R4 Strains with Selected Antibiotics
2.5. Cleavage of Plasmid DNA by Application with Fpg Glycosylases in Bacterial Cells
2.6. Cleavage of Plasmid DNA by Fpg Protein Modified by Selected Antibiotics
2.7. Statistical Analysis
3. Results
3.1. Chemistry
3.2. Toxicity of Tested Compounds
3.3. Modification of Plasmid DNA Isolated from E. coli R2–R4 Strains with Tested 1,2-Diarylethanoles
4. Discussion
5. Conclusions
- 1,2-diarylethanols with a specific structure are able to modify all strains of E. coli (R2–R4) and their plasmid DNA, then the spatial structure of LPS contained in their cell membrane is changed.
- The R4 type strain was the most sensitive amongst all the tested E. coli strains.
- The interaction of the analysed 1,2-diarylethanols with the cell membrane of the K12 strain shows differences in the O-antigen and the shortened oligosaccharide core compared to the analysed R-type strains, which may play an important role in the cellular response to the charged compounds.
- The toxicity of aromatic groups together with the alkyl substituents depends on their interaction with the membrane that may become involved in the structures of cell walls and change their hydrophobicity.
- Changes in the structure of the bacterial membrane and disturbances in its integrity may result in changes in the bacterial response to other biologically active compounds such as antibiotics.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Compliance with Ethical Standards
Abbreviations
| MIC | minimum inhibitory concentration |
| MBC | minimum bactericidal concentration |
| oc | open circle |
| ccc | covalently closed circle |
References
- Boyce, J.M. Alkohols as Surface Disinfectants in Healthcare Settings. Infect. Control. Hosp. Epidemiol 2018, 39, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Shimizu, K. Alcohol Denaturation: Thermodynamic Theory of Peptide Unit Solvation. J. Am. Chem. Soc. 1999, 121, 2387–2394. [Google Scholar] [CrossRef]
- Yano, T.; Miyahara, Y.; Morii, N.; Okano, T.; Kubota, H. Pentanol and Benzyl Alcohol Attack Bacterial Surface Structures Differently. Appl. Environ. Microbiol. 2015, 82, 402–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, D.D.M.; Lopes, L.K.D.O.; Hu, H.; Tipple, A.F.V.; Vickery, K. Alcohol fixation of bacteria to surgical instruments increases cleaning difficulty and may contribute to sterilization inefficacy. Am. J. Infect. Control. 2017, 45, e81–e86. [Google Scholar] [CrossRef]
- Keen, J.N.; Austin, M.; Huang, L.-S.; Messing, S.; Wyatt, J.D. Efficacy of Soaking in 70% Isopropyl Alcohol on Aerobic Bacterial Decontamination of Surgical Instruments and Gloves for Serial Mouse Laparotomies. J. Am. Assoc. Lab. Anim. Sci. 2010, 49, 832–837. [Google Scholar] [PubMed]
- Guenezan, J.; Drugeon, B.; O’Neill, R.; Caillaud, D.; Sénamaud, C.; Pouzet, C.; Seguin, S.; Frasca, D.; Mimoz, O. Skin antisepsis with chlorhexidine–alcohol versus povidone iodine–alcohol, combined or not with use of a bundle of new devices, for prevention of short-term peripheral venous catheter-related infectious complications and catheter failure: An open-label, single-centre, randomised, four-parallel group, two-by-two factorial trial: CLEAN 3 protocol study. BMJ Open 2019, 9, e028549. [Google Scholar] [CrossRef]
- Muñoz-Figueroa, G.P.; Ojo, O. The effectiveness of alcohol-based gel for hand sanitising in infection control. Br. J. Nurs. 2018, 27, 382–388. [Google Scholar] [CrossRef]
- Suchomel, M.; Eggers, M.; Maier, S.; Kramer, A.; Dancer, S.J.; Pittet, D. Evaluation of World Health Organization–Recommended Hand Hygiene Formulations. Emerg. Infect. Dis. 2020, 26, 2064–2068. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Padial, N.M.; Kingston, C.; Vantourout, J.C.; Schmitt, D.C.; Edwards, J.T.; Kruszyk, M.M.; Merchant, R.R.; Mykhailiuk, P.K.; Sanchez, B.B.; et al. A Radical Approach to Anionic Chemistry: Synthesis of Ketones, Alcohols, and Amines. J. Am. Chem. Soc. 2019, 141, 6726–6739. [Google Scholar] [CrossRef] [PubMed]
- Madduluri, V.K.; Sah, A.K. Metal complexes of 4,6-O-ethylidene-β-d-glucopyranosylamine derivatives and their application in organic synthesis. Carbohydr. Res. 2019, 485, 107798. [Google Scholar] [CrossRef]
- Nainwal, L.M.; Alam, M.M.; Shaquiquzzaman, M.; Marella, A.; Kamal, A. Combretastatin-based compounds with therapeutic characteristics: A patent review. Expert Opin. Ther. Patents 2019, 29, 703–731. [Google Scholar] [CrossRef]
- Hura, N.; Sawant, A.V.; Kumari, A.; Guchhait, S.K.; Panda, D. Combretastatin-Inspired Heterocycles as Antitubulin Anticancer Agents. ACS Omega 2018, 3, 9754–9769. [Google Scholar] [CrossRef]
- Nik, M.E.; Momtazi-Borojeni, A.A.; Zamani, P.; Navashenaq, J.G.; Iranshahi, M.; Jaafari, M.R.; Malaekeh-Nikouei, B. Targeted-nanoliposomal combretastatin A4 (CA-4) as an efficient antivascular candidate in the metastatic cancer treatment. J. Cell. Physiol. 2019, 234, 14721–14733. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Tang, Z.-H.; Sun, L.; Yu, H.-Y.; Li, J.; Zhao, M.-H.; Xu, H. Combretastatin A4/poly(L-glutamic acid)-graft-PEG conjugates self-assembled to nanoparticles. Asian J. Pharm. Sci. 2018, 13, 191–196. [Google Scholar] [CrossRef]
- Sherbet, G.V. Combretastatin analogues in cancer biology: A prospective view. J. Cell. Biochem. 2019, 121, 2127–2138. [Google Scholar] [CrossRef]
- Abma, E.; Daminet, S.; Smets, P.; Ni, Y.; De Rooster, H. Combretastatin A4-phosphate and its potential in veterinary oncology: A review. Veter. Comp. Oncol. 2015, 15, 184–193. [Google Scholar] [CrossRef] [Green Version]
- Kowalczyk, P.; Madej, A.; Paprocki, D.; Szymczak, M.; Ostaszewski, R. Coumarin Derivatives as New Toxic Compounds to Selected K12, R1–R4 E. coli Strains. Mateials 2020, 13, 2499. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Madej, A.; Szymczak, M.; Ostaszewski, R. α-Amidoamids as New Replacements of Antibiotics—Research on the Chosen K12, R2–R4 E. coli Strains. Mateials 2020, 13, 5169. [Google Scholar] [CrossRef] [PubMed]
- Mi, X.; Wang, C.; Huang, M.; Wu, Y.; Wu, Y. Silver-catalyzed carbonphosphonation of α,α-diaryl allylic alcohols: Synthesis of β-aryl-γ-ketophosphonates. Org. Biomol. Chem. 2014, 12, 8394–8397. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Han, X.; He, M.; Xiao, Y.; Qin, Z. Synthesis and fungicidal activity of 1,1-diaryl tertiary alcohols. Bioorganic Med. Chem. Lett. 2016, 26, 5936–5942. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Borkowski, A.; Czerwonka, G.; Cłapa, T.; Cieśla, J.; Misiewicz, A.; Borowiec, M.; Szala, M. The microbial toxicity of quaternary ammonium ionic liquids is dependent on the type of lipopolysaccharide. J. Mol. Liq. 2018, 266, 540–547. [Google Scholar] [CrossRef]
- Borkowski, A.; Kowalczyk, P.; Czerwonka, G.; Cie’sla, J.; Cłapa, T.; Misiewicz, A.; Szala, M.; Drabik, M. Interaction of quaternary ammonium ionic liquids with bacterial membranes—Studies with Escherichia coli R1–R4-type lipopolysaccharides. J. Mol. Liq. 2017, 246, 282–289. [Google Scholar] [CrossRef]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of Core Oligosaccharide Types in Lipopolysaccharides from Escherichia coli. Infect. Immun. 2000, 68, 1116–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciejewska, A.; Kaszowska, M.; Jachymek, W.; Lugowski, C.; Lukasiewicz, J. Lipopolysaccharide-linked Enterobacterial Common Antigen (ECALPS) Occurs in Rough Strains of Escherichia coli R1, R2, and R4. Int. J. Mol. Sci. 2020, 21, 6038. [Google Scholar] [CrossRef]
- Orlov, N.V.; Ananikov, V.P. NMR analysis of chiral alcohols and amines: Development of an environmentally benign “in tube” procedure with high efficiency and improved detection limit. Green Chem. 2011, 13, 1735–1744. [Google Scholar] [CrossRef]
- Pieczykolan, M.; Narczyk, A.; Stecko, S. The Synthesis of Chiral β,β-Diaryl Allylic Alcohols and Their Use in the Preparation of α-Tertiary Allylamines and Quaternary α-Amino Acids. J. Org. Chem. 2017, 82, 5636–5651. [Google Scholar] [CrossRef] [PubMed]
- Narczyk, A.; Stecko, S. The synthesis of unnatural α-alkyl- and α-aryl-substituted serine derivatives. Org. Biomol. Chem. 2020, 18, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Wink, D.A.; Laval, J. The Fpg protein, a DNA repair enzyme, is inhibited by the biomediator nitric oxide in vitro and in vivo. Carcinogenisis 1994, 15, 2125–2129. [Google Scholar] [CrossRef]
- Spellberg, B.; Chambers, H.F.; Musher, D.M.; Walsh, T.L.; Bayer, A.S. Evaluation of a Paradigm Shift From Intravenous Antibiotics to Oral Step-Down Therapy for the Treatment of Infective Endocarditis. JAMA Intern. Med. 2020, 180, 769–777. [Google Scholar] [CrossRef]
- Hong, J.; Ensom, M.H.H.; Lau, T.T.Y. What Is the Evidence for Co-trimoxazole, Clindamycin, Doxycycline, and Minocycline in the Treatment of Methicillin-Resistant Staphylococcus aureus (MRSA) Pneumonia? Ann. Pharmacother. 2019, 53, 1153–1161. [Google Scholar] [CrossRef]
- Sandoval, A.; Cofré, F.; Delpiano, L.; Izquierdo, G.; Labraña, Y.; Reyes, A. Reposicionando la cloxacilina como antibioticoterapia empírica inicial de la sepsis tardía neonatal. Rev. Chil. Infectol. 2015, 32, 182–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møller, P.; Jantzen, K.; Løhr, M.; Andersen, M.H.; Jensen, D.M.; Roursgaard, M.; Danielsen, P.H.; Jensen, A.; Loft, S. Searching for assay controls for the Fpg- and hOGG1-modified comet assay. Mutagenesis 2017, 33, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Dang, W.; Tong, C.; Yang, Y.; Liu, Y.; Liu, B.; Zhou, H.; Wang, W. A cascade amplification platform assisted with DNAzyme for activity analysis, kinetic study and effector screening of Fpg in vitro. Analyst 2019, 144, 1731–1740. [Google Scholar] [CrossRef]
- Massa, S.; Di Santo, R.; Retico, A.; Artico, M.; Simonetti, N.; Fabrizi, G.; Lamba, D. Antifungal agents. 1. Synthesis and antifungal activities of estrogen-like imidazole and triazole derivatives. Eur. J. Med. Chem. 1992, 27, 495–502. [Google Scholar] [CrossRef]
- Roche, M.; Lacroix, C.; Khoumeri, O.; Franco, D.; Neyts, J.; Terme, T.; Leyssen, P.; Vanelle, P. Synthesis, biological activity and structure–activity relationship of 4,5-dimethoxybenzene derivatives inhibitor of rhinovirus 14 infection. Eur. J. Med. Chem. 2014, 76, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Firth-Clark, S.; Willems, H.M.G.; Williams, A.; Harris, W. Generation and Selection of Novel Estrogen Receptor Ligands Using theDe NovoStructure-Based Design Tool, SkelGen. J. Chem. Inf. Model. 2006, 46, 642–647. [Google Scholar] [CrossRef]
- Saldan, I.; Semenyuk, Y.; Marchuk, I.; Reshetnyak, O. Chemical synthesis and application of palladium nanoparticles. J. Mater. Sci. 2015, 50, 2337–2354. [Google Scholar] [CrossRef]
- Girard, C.; Fayolle, K.; Kerros, S.; Leriche, F. Flow cytometric assessment of the antimicrobial properties of an essential oil mixture against Escherichia coli. J. Anim. Feed. Sci. 2019, 28, 187–198. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Yang, Y.; Zhang, P.; Du, Z.; Wang, C. Alkene Oxyalkylation Enabled by Merging Rhenium Catalysis with Hypervalent Iodine(III) Reagents via Decarboxylation. J. Am. Chem. Soc. 2013, 135, 18048–18051. [Google Scholar] [CrossRef]
- Weidmann, N.; Harenberg, J.H.; Knochel, P. Continuous Flow Preparation of (Hetero)benzylic Lithiums via Iodine–Lithium Exchange Reaction under Barbier Conditions. Org. Lett. 2020, 22, 5895–5899. [Google Scholar] [CrossRef]
- Karatoprak, G.Ş.; Akkol, E.K.; Genç, Y.; Bardakci, H.; Yücel, Ç.; Sobarzo-Sánchez, E. Combretastatins: An Overview of Structure, Probable Mechanisms of Action and Potential Applications. Molecules 2020, 25, 2560. [Google Scholar] [CrossRef] [PubMed]









| Compound Symbol | R1 | R2 | R3 | Yield 1 (%) |
|---|---|---|---|---|
| 4a | H | H | Me | 34 |
| 4b | Me | H | Me | 19 |
| 4c | CF3 | H | Me | 2 |
| 4d | Cl | H | Me | 34 |
| 4e | Br | H | Me | 6 |
| 4f | I | - | Me | 0(22) |
| 4h | -CHO | H | Me | 4 |
| 4i | -CH2OH | H | Me | 5 |
| 4j | CH3O- | H | Me | 1 |
| 4k | CH3O- | CH3O- | Me | 33 |
| 4l | Me3COC(O)NH- | H | Me | 9 |
| 4la | - | H | Me | 0(62) |
| 4ck | CF3-, CH3O- | CF3-, CH3O- | Me | 2 |
| 4q | H | H | Ph | 22 |
| 4t | H | H | Cl | 7 |
| 4u | H | H | naproxen | 0(212) |
| No of Samples | 4a | b | c | d | e | f | h | i | j | k | l | ł | ck | q | t | u | 7a | 7k | Type of Test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K12 | *** | *** | ** | ** | *** | ** | *** | *** | ** | MIC | |||||||||
| R2 | *** | *** | ** | ** | *** | ** | *** | *** | ** | MIC | |||||||||
| R3 | *** | *** | ** | ** | *** | ** | *** | *** | ** | MIC | |||||||||
| R4 | *** | *** | ** | ** | *** | ** | *** | *** | ** | MIC | |||||||||
| K12 | ** | ** | *** | *** | ** | *** | ** | *** | *** | MBC | |||||||||
| R2 | ** | ** | *** | *** | ** | *** | ** | *** | *** | MBC | |||||||||
| R3 | ** | ** | *** | *** | ** | *** | ** | *** | *** | MBC | |||||||||
| R4 | ** | ** | *** | *** | ** | *** | ** | *** | *** | MBC | |||||||||
| K12 | * | * | ** | ** | * | ** | * | *** | * | MBC/MIC | |||||||||
| R2 | * | * | ** | ** | * | ** | * | *** | * | MBC/MIC | |||||||||
| R3 | * | * | ** | ** | * | ** | * | *** | * | MBC/MIC | |||||||||
| R4 | * | * | ** | ** | * | ** | * | *** | * | MBC/MIC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalczyk, P.; Trzepizur, D.; Szymczak, M.; Skiba, G.; Kramkowski, K.; Ostaszewski, R. 1,2-Diarylethanols—A New Class of Compounds That Are Toxic to E. coli K12, R2–R4 Strains. Materials 2021, 14, 1025. https://doi.org/10.3390/ma14041025
Kowalczyk P, Trzepizur D, Szymczak M, Skiba G, Kramkowski K, Ostaszewski R. 1,2-Diarylethanols—A New Class of Compounds That Are Toxic to E. coli K12, R2–R4 Strains. Materials. 2021; 14(4):1025. https://doi.org/10.3390/ma14041025
Chicago/Turabian StyleKowalczyk, Paweł, Damian Trzepizur, Mateusz Szymczak, Grzegorz Skiba, Karol Kramkowski, and Ryszard Ostaszewski. 2021. "1,2-Diarylethanols—A New Class of Compounds That Are Toxic to E. coli K12, R2–R4 Strains" Materials 14, no. 4: 1025. https://doi.org/10.3390/ma14041025
APA StyleKowalczyk, P., Trzepizur, D., Szymczak, M., Skiba, G., Kramkowski, K., & Ostaszewski, R. (2021). 1,2-Diarylethanols—A New Class of Compounds That Are Toxic to E. coli K12, R2–R4 Strains. Materials, 14(4), 1025. https://doi.org/10.3390/ma14041025