A Higher Lignin Content in ugt72b37 Poplar Mutants Indicates a Role of Monolignol Glycosylation in Xylem Lignification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Characterization
2.2. CRISPR/Cas9 sgRNA Design and Vector Construction
2.3. Plant Material, Transformation, Mutation Screening, and Growth Conditions
2.4. Microscopy
2.5. Cell Wall Analysis, Lignin Composition, and Saccharification
2.6. Gene Expression Analysis
3. Results
3.1. UGT72B37 Protein Characterization
3.2. Generation of ugt72b37 Mutants via CRISPR/Cas9
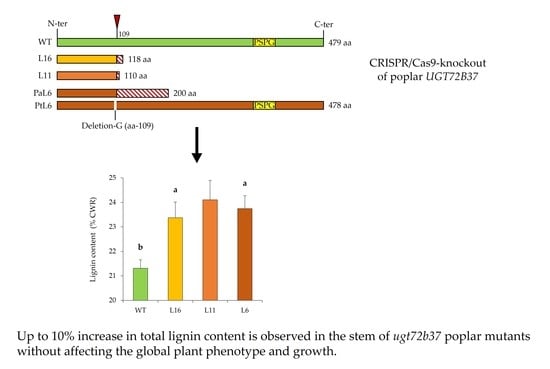
3.3. The ugt72b37 Mutation Triggers an Increased Lignin Content in Stem Xylem
3.4. Expression Level of Genes Involved in Monolignol Biosynthesis and Polymerization
3.5. Impact of the ugt72b37 Mutation on the Expression of the Other Poplar UGT72B Genes
4. Discussion
4.1. UGT72B37 Has a Role in the Xylem Lignification Process
4.2. The UGT72B37 Mutation in Poplar Has a Different Impact Than the UGT72B1 Mutation in Arabidopsis and the LuUGT175 Mutation in Flax
4.3. The Increase in Lignin Content Measured in the ugt72b37 Mutants May Be Linked to an Upregulation of CCR2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The Cell Biology of Lignification in Higher Plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef] [Green Version]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Cui, D.; Ye, Z.-H. Secondary Cell Wall Biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, R.A.; Barros, J. Lignin Biosynthesis: Old Roads Revisited and New Roads Explored. Open Biol. 2019, 9, 190215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.P.; Matthews, M.L.; Naik, P.P.; Williams, C.M.; Ducoste, J.J.; Sederoff, R.R.; Chiang, V.L. Flux Modeling for Monolignol Biosynthesis. Curr. Opin. Biotechnol. 2019, 56, 187–192. [Google Scholar] [CrossRef]
- Wang, J.P.; Liu, B.; Sun, Y.; Chiang, V.L.; Sederoff, R.R. Enzyme-Enzyme Interactions in Monolignol Biosynthesis. Front. Plant Sci. 2019, 9, 1942. [Google Scholar] [CrossRef] [Green Version]
- Xin, A.; Herburger, K. Mini Review: Transport of Hydrophobic Polymers Into the Plant Apoplast. Front. Plant Sci. 2021, 11, 590990. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Dixon, R.A.; Chen, F.; Mansfield, S.D.; Boerjan, W.; Ralph, J.; Crowley, M.F.; Beckham, G.T. Passive Membrane Transport of Lignin-Related Compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 23117–23123. [Google Scholar] [CrossRef]
- Perkins, M.; Smith, R.A.; Samuels, L. The Transport of Monomers during Lignification in Plants: Anything Goes but How? Curr. Opin. Biotechnol. 2019, 56, 69–74. [Google Scholar] [CrossRef]
- Perkins, M.L.; Schuetz, M.; Unda, F.; Chen, K.T.; Bally, M.B.; Kulkarni, J.A.; Yan, Y.; Pico, J.; Castellarin, S.D.; Mansfield, S.D.; et al. Monolignol Export by Diffusion down a Polymerization-Induced Concentration Gradient. Plant Cell 2022, 34, 2080–2095. [Google Scholar] [CrossRef]
- Ralph, J.; Brunow, G.; Harris, P.J.; Dixon, R.A.; Schatz, P.F.; Boerjan, W. Lignification: Are Lignins Biosynthesized via Simple Combinatorial Chemistry or via Proteinaceous Control and Template Replication? In Proceedings of the Recent Advances in Polyphenol Research; Wiley: Hoboken, NJ, USA, 2008; Volume 1, pp. 36–66. [Google Scholar]
- Tobimatsu, Y.; Schuetz, M. Lignin Polymerization: How Do Plants Manage the Chemistry so Well? Curr. Opin. Biotechnol. 2019, 56, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Väisänen, E.E.; Smeds, A.I.; Fagerstedt, K.V.; Teeri, T.H.; Willför, S.M.; Kärkönen, A. Coniferyl Alcohol Hinders the Growth of Tobacco BY-2 Cells and Nicotiana Benthamiana Seedlings. Planta 2015, 242, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Dima, O.; Morreel, K.; Vanholme, B.; Kim, H.; Ralph, J.; Boerjan, W. Small Glycosylated Lignin Oligomers Are Stored in Arabidopsis Leaf Vacuoles. Plant Cell 2015, 27, 695–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuyama, T.; Kawai, R.; Shitan, N.; Matoh, T.; Sugiyama, J.; Yoshinaga, A.; Takabe, K.; Fujita, M.; Yazaki, K. Proton-Dependent Coniferin Transport, a Common Major Transport Event in Differentiating Xylem Tissue of Woody Plants. Plant Physiol. 2013, 162, 918–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Väisänen, E.; Takahashi, J.; Obudulu, O.; Bygdell, J.; Karhunen, P.; Blokhina, O.; Laitinen, T.; Teeri, T.H.; Wingsle, G.; Fagerstedt, K.V.; et al. Hunting Monolignol Transporters: Membrane Proteomics and Biochemical Transport Assays with Membrane Vesicles of Norway Spruce. J. Exp. Bot. 2020, 71, 6379–6395. [Google Scholar] [CrossRef] [PubMed]
- Tsuyama, T.; Matsushita, Y.; Fukushima, K.; Takabe, K.; Yazaki, K.; Kamei, I. Proton Gradient-Dependent Transport of p-Glucocoumaryl Alcohol in Differentiating Xylem of Woody Plants. Sci. Rep. 2019, 9, 8900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.-C.; Liu, C.-J. ATP-Binding Cassette-like Transporters Are Involved in the Transport of Lignin Precursors across Plasma and Vacuolar Membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 22728–22733. [Google Scholar] [CrossRef] [Green Version]
- Shimada, N.; Munekata, N.; Tsuyama, T.; Matsushita, Y.; Fukushima, K.; Kijidani, Y.; Takabe, K.; Yazaki, K.; Kamei, I. Active Transport of Lignin Precursors into Membrane Vesicles from Lignifying Tissues of Bamboo. Plants 2021, 10, 2237. [Google Scholar] [CrossRef]
- Aoki, D.; Okumura, W.; Akita, T.; Matsushita, Y.; Yoshida, M.; Sano, Y.; Fukushima, K. Microscopic Distribution of Syringin in Freeze-fixed Syringa Vulgaris Stems. Plant Direct 2019, 3, e00155. [Google Scholar] [CrossRef]
- Aoki, D.; Hanaya, Y.; Akita, T.; Matsushita, Y.; Yoshida, M.; Kuroda, K.; Yagami, S.; Takama, R.; Fukushima, K. Distribution of Coniferin in Freeze-Fixed Stem of Ginkgo Biloba L. by Cryo-TOF-SIMS/SEM. Sci. Rep. 2016, 6, 31525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freudenberg, K. Biosynthesis and Constitution of Lignin. Nature 1959, 183, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Freudenberg, K.; Harkin, J.M. The Glucosides of Cambial Sap of Spruce. Phytochemistry 1963, 2, 189–193. [Google Scholar] [CrossRef]
- Fukushima, K. Distribution and Seasonal Changes of Monolignol Glucosides in Pinus Thunbergii. J. Jap. Wood Res. Soc. 1997, 43, 254–259. [Google Scholar]
- Tsuyama, T.; Takabe, K. Distribution of Lignin and Lignin Precursors in Differentiating Xylem of Japanese Cypress and Poplar. J. Wood Sci. 2014, 60, 353–361. [Google Scholar] [CrossRef]
- Yoshinaga, A.; Kamitakahara, H.; Takabe, K. Distribution of Coniferin in Differentiating Normal and Compression Woods Using MALDI Mass Spectrometric Imaging Coupled with Osmium Tetroxide Vapor Treatment. Tree Physiol. 2016, 36, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Dharmawardhana, D.P.; Ellis, B.E.; Carlson, J.E. A Beta-Glucosidase from Lodgepole Pine Xylem Specific for the Lignin Precursor Coniferin. Plant Physiol. 1995, 107, 331–339. [Google Scholar] [CrossRef]
- Escamilla-Treviño, L.L.; Chen, W.; Card, M.L.; Shih, M.-C.; Cheng, C.-L.; Poulton, J.E. Arabidopsis Thaliana Beta-Glucosidases BGLU45 and BGLU46 Hydrolyse Monolignol Glucosides. Phytochemistry 2006, 67, 1651–1660. [Google Scholar] [CrossRef]
- Rolando, C.; Daubresse, N.; Pollet, B.; Jouanin, L.; Lapierre, C. Lignification in Poplar Plantlets Fed with Deuterium-Labelled Lignin Precursors. Comptes Rendus Biol. 2004, 327, 799–807. [Google Scholar] [CrossRef]
- Terashima, N. Non-Destructive Approaches to Identify the Ultrastructure of Lignified Ginkgo Cell Walls. Int. J. Plant Develop. Biol. 2007, 1, 170–177. [Google Scholar]
- Terashima, N.; Ko, C.; Matsushita, Y.; Westermark, U. Monolignol Glucosides as Intermediate Compounds in Lignin Biosynthesis. Revisiting the Cell Wall Lignification and New 13C-Tracer Experiments with Ginkgo Biloba and Magnolialiliiflora. Holzforschung 2016, 70, 801–810. [Google Scholar] [CrossRef]
- Terashima, N.; Fukushima, K.; Sano, Y.; Takabe, K. Heterogeneity in Formation of Lignin. X. Visualization of Lignification Process in Differentiating Xylem of Pine by Microautoradiography. Holzforschung 1988, 42, 347–350. [Google Scholar] [CrossRef]
- Tsuji, Y.; Chen, F.; Yasuda, S.; Fukushima, K. Unexpected Behavior of Coniferin in Lignin Biosynthesis of Ginkgo Biloba L. Planta 2005, 222, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, Y.; Fukushima, K. Behavior of Monolignol Glucosides in Angiosperms. J. Agric. Food Chem. 2004, 52, 7651–7659. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, Y.; Tobimatsu, Y.; Lam, P.Y.; Mizukami, T.; Sakurai, S.; Kamitakahara, H.; Takano, T. Possible Mechanisms for the Generation of Phenyl Glycoside-Type Lignin–Carbohydrate Linkages in Lignification with Monolignol Glucosides. Plant J. 2020, 104, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Lanot, A.; Hodge, D.; Lim, E.-K.; Vaistij, F.E.; Bowles, D.J. Redirection of Flux through the Phenylpropanoid Pathway by Increased Glucosylation of Soluble Intermediates. Planta 2008, 228, 609–616. [Google Scholar] [CrossRef]
- Lanot, A.; Hodge, D.; Jackson, R.G.; George, G.L.; Elias, L.; Lim, E.-K.; Vaistij, F.E.; Bowles, D.J. The Glucosyltransferase UGT72E2 Is Responsible for Monolignol 4-O-Glucoside Production in Arabidopsis Thaliana. Plant J. 2006, 48, 286–295. [Google Scholar] [CrossRef]
- Lim, E.-K.; Jackson, R.G.; Bowles, D.J. Identification and Characterisation of Arabidopsis Glycosyltransferases Capable of Glucosylating Coniferyl Aldehyde and Sinapyl Aldehyde. FEBS Lett. 2005, 579, 2802–2806. [Google Scholar] [CrossRef] [Green Version]
- Lim, E.-K.; Li, Y.; Parr, A.; Jackson, R.; Ashford, D.A.; Bowles, D.J. Identification of Glucosyltransferase Genes Involved in Sinapate Metabolism and Lignin Synthesis in Arabidopsis. J. Biol. Chem. 2001, 276, 4344–4349. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.-S.; Huang, X.-X.; Li, Q.; Cao, Y.; Bao, Y.; Meng, X.-F.; Li, Y.-J.; Fu, C.; Hou, B.-K. UDP-Glycosyltransferase 72B1 Catalyzes the Glucose Conjugation of Monolignols and Is Essential for the Normal Cell Wall Lignification in Arabidopsis Thaliana. Plant J. 2016, 88, 26–42. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhu, W.; Shan, X.; Liu, J.; Zhao, L.; Zhao, Q. Glycoside-Specific Metabolomics Combined with Precursor Isotopic Labeling for Characterizing Plant Glycosyltransferases. Mol. Plant 2022, 15, 1517–1532. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Dai, X.; Gao, L.; Guo, L.; Zhuang, J.; Liu, Y.; Ma, X.; Wang, R.; Xia, T.; Wang, Y. Functional Analysis of an Uridine Diphosphate Glycosyltransferase Involved in the Biosynthesis of Polyphenolic Glucoside in Tea Plants (Camellia Sinensis). J. Agric. Food Chem. 2017, 65, 10993–11001. [Google Scholar] [CrossRef]
- Louveau, T.; Leitao, C.; Green, S.; Hamiaux, C.; van der Rest, B.; Dechy-Cabaret, O.; Atkinson, R.G.; Chervin, C. Predicting the Substrate Specificity of a Glycosyltransferase Implicated in the Production of Phenolic Volatiles in Tomato Fruit. FEBS J. 2011, 278, 390–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallage, N.J.; Hansen, E.H.; Kannangara, R.; Olsen, C.E.; Motawia, M.S.; Jørgensen, K.; Holme, I.; Hebelstrup, K.; Grisoni, M.; Møller, B.L. Vanillin Formation from Ferulic Acid in Vanilla Planifolia Is Catalysed by a Single Enzyme. Nat Commun. 2014, 5, 4037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Yang, J.; Jiang, Y.; Wang, J.; Liu, Y.; Zhao, Y.; Jin, B.; Wang, X.; Chen, T.; Kang, L.; et al. Functional Characterization of UDP-Glycosyltransferases Involved in Anti-Viral Lignan Glycosides Biosynthesis in Isatis Indigotica. Front. Plant Sci. 2022, 13, 921815. [Google Scholar] [CrossRef] [PubMed]
- Härtl, K.; Huang, F.-C.; Giri, A.P.; Franz-Oberdorf, K.; Frotscher, J.; Shao, Y.; Hoffmann, T.; Schwab, W. Glucosylation of Smoke-Derived Volatiles in Grapevine (Vitis Vinifera) Is Catalyzed by a Promiscuous Resveratrol/Guaiacol Glucosyltransferase. J. Agric. Food Chem. 2017, 65, 5681–5689. [Google Scholar] [CrossRef]
- Wang, H.; Feng, X.; Zhang, Y.; Wei, D.; Zhang, Y.; Jin, Q.; Cai, Y. PbUGT72AJ2-Mediated Glycosylation Plays an Important Role in Lignin Formation and Stone Cell Development in Pears (Pyrus Bretschneideri). Int. J. Mol. Sci. 2022, 23, 7893. [Google Scholar] [CrossRef]
- Speeckaert, N.; Adamou, N.M.; Hassane, H.A.; Baldacci-Cresp, F.; Mol, A.; Goeminne, G.; Boerjan, W.; Duez, P.; Hawkins, S.; Neutelings, G.; et al. Characterization of the UDP-Glycosyltransferase UGT72 Family in Poplar and Identification of Genes Involved in the Glycosylation of Monolignols. Int. J. Mol. Sci. 2020, 21, 5018. [Google Scholar] [CrossRef]
- Speeckaert, N.; El Jaziri, M.; Baucher, M.; Behr, M. UGT72, a Major Glycosyltransferase Family for Flavonoid and Monolignol Homeostasis in Plants. Biology 2022, 11, 441. [Google Scholar] [CrossRef]
- Xie, D.; Yang, X.; He, R.; Huo, H.; Ye, Z.; Ren, X.; Yuan, H.; Dai, Z.; Sun, J.; Su, J. Comprehensive Analysis of the UDP-glycosyltransferase Gene Family in Flax [Linum Usitatissimum L.] and Functional Verification of the Role of LuUGT175 in the Regulation of Lignin Biosynthesis. Ind. Crops Prod. 2022, 188, 115720. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, LLC. The PyMOL Molecular Graphics System; Version 1.8; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-MPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 Web Portal for Protein Modelling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic Identification of Cell Cycle-Dependent Yeast Nucleocytoplasmic Shuttling Proteins by Prediction of Composite Motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [Green Version]
- Heigwer, F.; Kerr, G.; Boutros, M. E-CRISP: Fast CRISPR Target Site Identification. Nat. Methods 2014, 11, 122–123. [Google Scholar] [CrossRef]
- Jacobs, T.B.; Martin, G.B. High-Throughput CRISPR Vector Construction and Characterization of DNA Modifications by Generation of Tomato Hairy Roots. JoVE 2016, 110, e53843. [Google Scholar] [CrossRef] [Green Version]
- Leple, J.C.; Brasileiro, A.C.M.; Michel, M.F.; Delmotte, F.; Jouanin, L. Transgenic Poplars: Expression of Chimeric Genes Using Four Different Constructs. Plant Cell Rep. 1992, 11, 137–141. [Google Scholar] [CrossRef]
- Lu, F.; Wang, C.; Chen, M.; Yue, F.; Ralph, J. A Facile Spectroscopic Method for Measuring Lignin Content in Lignocellulosic Biomass. Green Chem. 2021, 23, 5106–5112. [Google Scholar] [CrossRef]
- Méchin, V.; Laluc, A.; Legée, F.; Cézard, L.; Denoue, D.; Barrière, Y.; Lapierre, C. Impact of the Brown-Midrib Bm5 Mutation on Maize Lignins. J. Agric. Food Chem. 2014, 62, 5102–5107. [Google Scholar] [CrossRef] [PubMed]
- Pettengill, E.A.; Parmentier-Line, C.; Coleman, G.D. Evaluation of QPCR Reference Genes in Two Genotypes of Populus for Use in Photoperiod and Low-Temperature Studies. BMC Res. Notes 2012, 5, 366. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. QBase Relative Quantification Framework and Software for Management and Automated Analysis of Real-Time Quantitative PCR Data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paquette, S.; Møller, B.L.; Bak, S. On the Origin of Family 1 Plant Glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef]
- Baldacci-Cresp, F.; Le Roy, J.; Huss, B.; Lion, C.; Créach, A.; Spriet, C.; Duponchel, L.; Biot, C.; Baucher, M.; Hawkins, S.; et al. UDP-GLYCOSYLTRANSFERASE 72E3 Plays a Role in Lignification of Secondary Cell Walls in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 6094. [Google Scholar] [CrossRef]
- Behr, M.; El Jaziri, M.; Baucher, M. Glycobiology of the Plant Secondary Cell Wall Dynamics. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Ďurkovič, J.; Kaňuchová, A.; Kačík, F.; Mamoňová, M.; Lengyelová, A. Wood Ontogeny during the First Year of Hybrid Poplar Development. Biol. Plant 2013, 57, 591–596. [Google Scholar] [CrossRef]
- Orella, M.J.; Gani, T.Z.H.; Vermaas, J.V.; Stone, M.L.; Anderson, E.M.; Beckham, G.T.; Brushett, F.R.; Román-Leshkov, Y. Lignin-KMC: A Toolkit for Simulating Lignin Biosynthesis. ACS Sustain. Chem. Eng. 2019, 7, 18313–18322. [Google Scholar] [CrossRef]
- Umezawa, T. Lignin Modification in Planta for Valorization. Phytochem. Rev. 2018, 17, 1305–1327. [Google Scholar] [CrossRef]
- Wei, K.; Zhao, Y.; Zhou, H.; Jiang, C.; Zhang, B.; Zhou, Y.; Song, X.; Lu, M. PagMYB216 Is Involved in the Regulation of Cellulose Synthesis in Populus. Mol. Breeding 2019, 39, 65. [Google Scholar] [CrossRef]
- Qin, S.; Fan, C.; Li, X.; Li, Y.; Hu, J.; Li, C.; Luo, K. LACCASE14 Is Required for the Deposition of Guaiacyl Lignin and Affects Cell Wall Digestibility in Poplar. Biotechnol. Biofuels 2020, 13, 197. [Google Scholar] [CrossRef]
- Koshiba, T.; Yamamoto, N.; Tobimatsu, Y.; Yamamura, M.; Suzuki, S.; Hattori, T.; Mukai, M.; Noda, S.; Shibata, D.; Sakamoto, M.; et al. MYB-Mediated Upregulation of Lignin Biosynthesis in Oryza Sativa towards Biomass Refinery. Plant Biotechnol. 2017, 34, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Meester, B.; Madariaga Calderón, B.; de Vries, L.; Pollier, J.; Goeminne, G.; Van Doorsselaere, J.; Chen, M.; Ralph, J.; Vanholme, R.; Boerjan, W. Tailoring Poplar Lignin without Yield Penalty by Combining a Null and Haploinsufficient CINNAMOYL-CoA REDUCTASE2 Allele. Nat. Commun. 2020, 11, 5020. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wu, J.; Lin, L.; Li, P.; Li, S.; Wang, Y.; Li, J.; Sun, Q.; Liang, J.; Wang, Y. Overexpression of Cinnamoyl-CoA Reductase 2 in Brassica Napus Increases Resistance to Sclerotinia Sclerotiorum by Affecting Lignin Biosynthesis. Front. Plant Sci. 2021, 12, 2045. [Google Scholar] [CrossRef] [PubMed]
- Cesarino, I. Structural Features and Regulation of Lignin Deposited upon Biotic and Abiotic Stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amadou Hassane, H.; Behr, M.; Guérin, C.; Sibout, R.; Mol, A.; Baragé, M.; El Jaziri, M.; Baucher, M. A Higher Lignin Content in ugt72b37 Poplar Mutants Indicates a Role of Monolignol Glycosylation in Xylem Lignification. Forests 2022, 13, 2167. https://doi.org/10.3390/f13122167
Amadou Hassane H, Behr M, Guérin C, Sibout R, Mol A, Baragé M, El Jaziri M, Baucher M. A Higher Lignin Content in ugt72b37 Poplar Mutants Indicates a Role of Monolignol Glycosylation in Xylem Lignification. Forests. 2022; 13(12):2167. https://doi.org/10.3390/f13122167
Chicago/Turabian StyleAmadou Hassane, Hadjara, Marc Behr, Claire Guérin, Richard Sibout, Adeline Mol, Moussa Baragé, Mondher El Jaziri, and Marie Baucher. 2022. "A Higher Lignin Content in ugt72b37 Poplar Mutants Indicates a Role of Monolignol Glycosylation in Xylem Lignification" Forests 13, no. 12: 2167. https://doi.org/10.3390/f13122167
APA StyleAmadou Hassane, H., Behr, M., Guérin, C., Sibout, R., Mol, A., Baragé, M., El Jaziri, M., & Baucher, M. (2022). A Higher Lignin Content in ugt72b37 Poplar Mutants Indicates a Role of Monolignol Glycosylation in Xylem Lignification. Forests, 13(12), 2167. https://doi.org/10.3390/f13122167