Bacmid Expression of Granulovirus Enhancin En3 Accumulates in Cell Soluble Fraction to Potentiate Nucleopolyhedrovirus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects, Cells and Viruses
2.2. Recombinant BacPh and BacPhEn3 Virus DNAs
2.3. Transfection of Sf9 Cells with Recombinant BacPh and BacPhEn3 Virus DNAs
2.4. Production of BacPh and BacEn3 OBs in Larvae
2.5. Detection and Localization of En3 Protein
2.6. Insect Bioassays
3. Results
3.1. Recombinant BacPh and BacPhEn3 Virus DNAs
3.2. Transfection of Sf9 Cells with Recombinant BacPh and BacPhEn3 Virus DNAs
3.3. Production of BacPh and BacPhEn3 OBs in Larvae
3.4. Enhancin 3 Is Solubilized in the Cell Medium
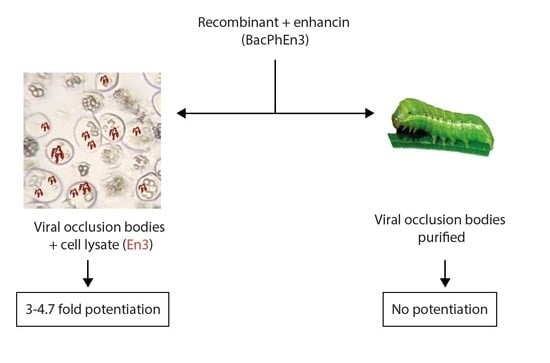
3.5. Biological Activity of BacPhEn3 OBs Produced in Cell Culture and in Larvae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Derksen, A.C.G.; Granados, R.R. Alteration of a lepidopteran peritrophic membrane by baculoviruses and enhancement of viral infectivity. Virology 1988, 167, 242–250. [Google Scholar] [CrossRef]
- Wang, M.; Wang, J.; Yin, F.; Tan, Y.; Deng, F.; Chen, X.; Jehle, J.A.; Vlak, J.M.; Hu, Z.; Wang, H. Unravelling the entry mechanism of baculoviruses and its evolutionary implications. J. Virol. 2014, 88, 2301–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Granados, R.R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. USA 1997, 94, 6977–6982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Granados, R.R. Molecular structure of the peritrophic membrane (PM): Identification of potential PM target sites for insect control. Arch. Insect Biochem. Physiol. 2001, 47, 110–118. [Google Scholar] [CrossRef]
- Wang, M.; Hu, Z. Cross-talking between baculoviruses and host insects towards a successful infection. Philos. Trans. R. Soc. B 2019, 374, 20180324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erlandson, M.A.; Toprak, U.; Hedegus, D.D. Role of the peritrophic matrix in insect-pathogen interactions. J. Insect Physiol. 2019, 117, 103894. [Google Scholar] [CrossRef]
- Gallo, L.G.; Corsaro, B.G.; Hughes, P.R.; Granados, R.R. In vivo enhancement of baculovirus infection by the viral enhancing factor of a granulosis virus of the cabbage looper Trichoplusia ni (Lepidoptera: Noctuidae). J. Invertebr. Pathol. 1991, 58, 203–210. [Google Scholar] [CrossRef]
- Gijzen, M.; Röelvink, P.; Granados, R. Characterization of viral enhancing activity from Trichoplusia ni granulosis virus. J. Invertebr. Pathol. 1995, 65, 289–294. [Google Scholar] [CrossRef]
- Guo, H.F.; Fang, J.C.; Wang, J.P.; Zhong, W.F.; Liu, B.S. Interaction of Xestia c-nigrum granulovirus with peritrophic matrix and Spodoptera litura nucleopolyhedrovirus in Spodoptera litura. J. Econ. Entomol. 2007, 100, 20–25. [Google Scholar] [CrossRef]
- Lepore, L.S.; Roelvink, P.R.; Granados, R.R. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J. Invertebr. Pathol. 1996, 68, 131–140. [Google Scholar] [CrossRef]
- Tanada, Y.; Himeno, M.; Omi, E.M. Isolation of a factor, from the capsule of a granulosis virus, synergistic for a nuclear-polyhedrosis virus of the armyworm. J. Invertebr. Pathol. 1973, 21, 31–90. [Google Scholar] [CrossRef]
- Wang, P.; Hammer, D.A.; Granados, R.R. Interaction of Trichoplusia ni granulosis virus-encoded enhancin with the midgut epithelium and peritrophic membrane of four lepidopteran insects. J. Gen. Virol. 1994, 75, 1961–1967. [Google Scholar] [CrossRef]
- Bischoff, D.S.; Slavicek, J.M. Molecular analysis of an enhancin gene in the Lymantria dispar nuclear polyhedrosis virus. J. Virol. 1997, 71, 8133–8140. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, L.; Moore, K.; Donly, C.; Theilmann, D.A.; Erlandson, M. Characterization of Mamestra configurata nucleopolyhedrovirus enhancin and its functional analysis via expression in an Autographa californica M nucleopolyhedrovirus recombinant. J. Gen. Virol. 2003, 84, 123–132. [Google Scholar] [CrossRef]
- Slavicek, J.M.; Popham, H.J.R. The Lymantria dispar nucleopolyhedrovirus enhancins are components of occlusion-derived virus. J. Virol. 2005, 79, 10578–10588. [Google Scholar] [CrossRef] [Green Version]
- Bivian-Hernández, M.A.; López-Tlacomulco, J.; Mares-Mares, E.; Ibarra, J.E.; Del Rincón-Castro, M.C. Genomic analysis of a Trichoplusia ni betabaculovirus (TnGV) with three different viral enhancing factors and two unique genes. Arch. Virol. 2017, 162, 3705–3715. [Google Scholar] [CrossRef]
- Tanada, Y.; Hess, R.T.; Omi, E.M. Invasion of a nuclear polyhedrosis virus in midgut of the armyworm, Pseudaletia unipuncta, and the enhancement of a synergistic enzyme. J. Invertebr. Pathol. 1975, 26, 99–104. [Google Scholar] [CrossRef]
- Hayakawa, T.; Shimojo, E.I.; Mori, M.; Kaido, M.; Furusawa, I.; Miyata, S.; Sano, Y.; Matsumoto, T.; Hashimoto, Y.; Granados, R. Enhancement of baculovirus infection in Spodoptera exigua (Lepidoptera: Noctuidae) larvae with Autographa californica nucleopolyhedrovirus or Nicotiana tabacum engineered with a granulovirus enhancin gene. Appl. Entomol. Zool. 2000, 35, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Del Rincón-Castro, M.C.; Ibarra, J.E. Effect of a nuclepolyhedrovirus of Autographa californica expressing the enhancin gene of Trichoplusia ni granulovirus on T. ni larvae. Biocontrol Sci. Technol. 2005, 15, 701–710. [Google Scholar] [CrossRef]
- Lei, C.; Yang, S.; Lei, W.; Nyamwasa, I.; Hu, J.; Sun, X. Displaying enhancing factors on the surface of occlusion bodies improves the insecticidal efficacy of a baculovirus. Pest Manag. Sci. 2020, 76, 1363–1370. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, L.; Ma, R.; Fang, W.; Hu, J.; Lei, C.; Sun, X. Improving baculovirus infectivity by efficiently embedding enhancing factors into occlusion bodies. Appl. Environ. Microbiol. 2017, 83, e00595-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemoto, Y.; Mitsuhashi, W.; Murakami, R.; Konishi, H.; Miyamato, K. The N-terminal region of an entomopoxvirus fusolin is essential for the enhancement of peroral infection, whereas the C-Terminal region is eliminated in digestive juice. J. Virol. 2008, 82, 12406–12415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, G.L.; Leppa, N.; Dickerson, W.A. Velvetbean caterpillar: A rearing procedure and artificial medium. J. Econ. Entomol. 1976, 69, 487–488. [Google Scholar] [CrossRef]
- King, L.A.; Possee, R.D. The Baculovirus Expression System. A Laboratory Guide; Chapman & Hall: London, UK, 1992. [Google Scholar]
- Ayres, M.D.; Howard, S.C.; Kuzio, J.; López-Ferber, M.; Possee, R.D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 1994, 202, 586–605. [Google Scholar] [CrossRef]
- Luckow, V.A.; Lee, C.; Barry, G.F.; Olins, P.O. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 1993, 67, 4566–4579. [Google Scholar] [CrossRef] [Green Version]
- Booth, W.T.; Schlachter, C.R.; Pote, S.; Ussin, N.; Mank, N.J.; Klapper, V.; Offermann, L.R.; Tang, C.; Hurlburt, B.K.; Chruszcz, M. Impact of an N-terminal polyhistidine Tag on protein thermal stability. ACS Omega 2018, 3, 760–768. [Google Scholar] [CrossRef]
- Galloway, C.S.; Wang, P.; Winstanley, D.; Jones, I.M. Comparison of the bacterial Enhancin-like proteins from Yersinia and Bacillus spp. with a baculovirus Enhancin. J. Invertebrt. Pathol. 2005, 90, 134–137. [Google Scholar] [CrossRef]
- O’Reilly, D.R.; Miller, L.K.; Luckow, V.A. Baculovirus Expression Vectors. A Laboratory Manual; Oxford University Press: New York, NY, USA, 1994. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Hughes, P.R.; Wood, H.A. A synchronous per oral technique for the bioassay of insect viruses. J. Invertebr. Pathol. 1981, 37, 154–159. [Google Scholar] [CrossRef]
- Jamovi. Jamovi Statistical Software v.1.2.27.0. Available online: https://www.jamovi.org (accessed on 15 September 2020).
- LeOra Software. POLO-PC: A User’s Guide to Probit or Logit Analysis; LeOra Software: Berkeley, CA, USA, 1987. [Google Scholar]
- Durantel, D.; Croizier, L.; Ayres, M.D.; Croizier, G.; Possee, R.D.; López-Ferber, M. The pnk/pnl gene (ORF 86) of Autographa californica nucleopolyhedrovirus is a nonessential, immediate early gene. J. Gen. Virol. 1998, 79, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Simón, O.; Williams, T.; López-Ferber, M.; Caballero, P. Genetic structure of a Spodoptera frugiperda nucleopolyhedrovirus population: High prevalence of deletion genotypes. Appl. Environ. Microbiol. 2004, 70, 5579–5588. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.X.; Liang, Z.P.; Chen, X.H.; Song, X.F.; Wang, L.W.; Shao, X.F. Expression and synergistic function of enhancin-like gene of Agrotis segetum granulovirus. Chin. J. Virol. 2012, 28, 258–264. [Google Scholar]
- Peng, J.; Zhong, J.; Granados, R.R. A baculovirus enhancin alters the permeability of a mucosal midgut peritrophic matrix from lepidopteran larvae. J. Insect Physiol. 1999, 45, 159–166. [Google Scholar] [CrossRef]
- Popham, H.J.R.; Bischoff, D.S.; Slavicek, J.M. Both Lymantria dispar nucleopolyhedrovirus enhancin genes contribute to viral potency. J. Virol. 2001, 75, 8639–8648. [Google Scholar] [CrossRef] [Green Version]
- Thiem, S.M. Baculovirus genes affecting host function. In Vitro Cell. Dev. Biol. Anim. 2009, 45, 111–126. [Google Scholar] [CrossRef]
- Toprak, U.; Harris, S.; Baldwin, D.; Theilmann, D.; Gillot, C.; Hedegus, D.D.; Erlandson, M.A. Role of enhancin in Mamestra configurata nucleopolyhedrovirus virulence: Selective degradation of host peritrophic matrix proteins. J. Gen. Virol. 2012, 93, 744–753. [Google Scholar] [CrossRef]
- Shi, X.; Chamankhah, M.; Visal-Shah, S.; Hemmingsen, S.M.; Erlandson, M.; Braun, L.; Alting-Mees, M.; Khachatourians, G.G.; O’Grady, M.; Hedegus, D.D. Modeling the structure of the Type I peritrophic matrix: Characterization of a Mamestra configurata intestinal mucin and a novel peritrophin containing 19 chitin binding domains. Insect Biochem. Mol. Biol. 2004, 34, 1101–1115. [Google Scholar] [CrossRef]
- Slavicek, J.M. Baculovirus enhancins and their role in viral pathogenicity. In Molecular Virology; Adoga, M.P., Ed.; InTech: Rijeka, Croatia, 2012; pp. 147–168. [Google Scholar]
- Je, Y.H.; Jin, B.B.; Park, H.W.; Roh, J.Y.; Chang, J.H.; Seo, S.J.; Olszewski, D.R.; O’Reilly, D.R.; Kang, S.K. Baculovirus expression vectors that incorporate the foreign protein into viral occlusion bodies. Biotechniques 2003, 34, 81–87. [Google Scholar] [CrossRef]
- Sun, X. History and current status of development and use of viral insecticides in China. Viruses 2015, 7, 306–319. [Google Scholar] [CrossRef]
- Tanada, Y. Synergism between two viruses of the armyworm, Pseudaletia unipuncta (Haworth) (Lepidoptera, Noctuidae). J. Insect Pathol. 1959, 1, 215–231. [Google Scholar]
- Biedma, M.E.; Salvador, R.; Ferrelli, M.L.; Sciocco-Cap, A.; Romanowski, V. Effect of the interaction between Anticarsia gemmatalis multiple nucleopolyhedrovirus and Epinotia aporema granulovirus, on Anticarsia gemmatalis (Lepidoptera: Noctuidae) larvae. Biol. Control 2015, 91, 17–21. [Google Scholar] [CrossRef]
- Jeyarani, S.; Karuppuchamy, P. Investigations on the enhancing efficacy of granulovirus on nucleopolyhedrovirus of Helicoverpa armigera (Hübner). J. Biopestic. 2010, 3, 172–176. [Google Scholar]
- Cao, J.; Ibrahim, H.; Garcia, J.J.; Mason, H.; Granados, R.R.; Earle, E.D. Transgenic tobacco plants carrying a baculovirus enhancin gene slow the development and increase the mortality of Trichoplusia ni larvae. Plant Cell Rep. 2002, 21, 244–250. [Google Scholar]
- Mori, M.; Kitamura, H.; Kondo, A.; Dohi, K.; Mori, M.; Kaido, M.; Mise, K.; Shimojyo, E.; Hashimoto, Y. Expression of an enhancin gene from the Trichoplusia ni granulosis virus confers resistance to lepidopterous insect pests to rice. Plant Biotechnol. 2006, 23, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Granados, R.R.; Fu, Y.; Corsaro, B.; Hughes, P.R. Enhancement of Bacillus thuringiensis toxicity to lepidopterous species with the enhancin from Trichoplusia ni granulovirus. Biol. Control 2001, 20, 153–159. [Google Scholar] [CrossRef]
- Guo, W.; Kain, W.; Wang, P. Effects of disruption of the peritrophic membrane on larval susceptibility to Bt toxin Cry1Ac in cabbage loopers. J. Insect Physiol. 2019, 117, 103897. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, Montreal, QC, Canada, 28 June–1 July 1998; Glasgow, J., Littlejohn, T., Major, F., Lathrop, R., Sankoff, D., Sensen, C., Eds.; AAAI Press: Menlo Park, CA, USA, 1998; pp. 175–182. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 19, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Primer | Sequence | Amplification Purpose |
|---|---|---|
| En3-TnGV-Fw | 5′-TCTCTAGAGCTGCATTAATTATAAGACTATGTC -3 | En3 amplification. TnGV DNA was used a template. Underlined—the XbaI site. Double underlined—En3 promoter and ATG start codon; nt 153,590 to 153,614 in the TnGV genome. Tm 51 °C. |
| En3-TnGV-Rv | 5′-CCCTGCAGTTAGAACGCTATCATTTTTAACG-3 | En3 amplification. TnGV DNA was used a template. Underlined—the PstI site. Double underlined —the en3 TAA stop codon; nt 156,293 to 156,315 in TnGV genome. Tm 50 °C. |
| Ph-Fw | 5′-CGCTCGAGGCCGGCATAGTACGC-3′ | Ph amplification. AcMNPV C6 DNA was used a template. Underlined—the XhoI site; nt 4197 to 4211 in AcMNPV genome. Tm 56 °C. |
| Ph-Rv | 5′-CGCCATGGTTAATACGCCGGACCAGTG-3′ | Ph amplification. AcMNPV C6 DNA was used a template. Underlined—the NcoI site. Double underlined—the ph TAA stop codon; nt 5239 to 5257 in AcMNPV genome. Tm 54 °C. |
| En3-Seq-Fw | 5′-CCGTACCCGCAAATATG-3′ | En3 sequence confirmation. Primer that annealed at nt 1053 to 1073 in the en3 gene. Tm 53 °C. |
| pFBD-seq-Fw | 5′-CCGTGTTTCAGTTAGCC-3′ | En3 sequence confirmation. Primer that annealed at nt 7563 to 7579 in pFBD plasmid. Tm 54 °C. |
| M13-Fw | 5’-CCCAGTCACGACGTTGTAAAACG-3´ | For confirmation of correct insertion of en3 and ph. Primer that flanked the mini att-Tn7 site of the bacmid. Tm 54 °C. |
| M13-Rv | 5´-AGCGGATAACAATTTCACACAGG-3´ | For confirmation of correct insertion of en3 and ph. Primer that flanked the mini att-Tn7 site of the bacmid. Tm 53 °C. |
| Instar | Virus | Slope ± SE | LC50 (OBs/mL) | 95% Conf. Interval | Relative | 95% Conf. Interval | ||
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Potency | Lower | Upper | ||||
| Second instar | OBs and cell lysate from Sf9 cells | |||||||
| BacPh | 0.306 ± 0.078 | 1.33 × 105 | 4.30 × 104 | 8.75 × 105 | 1 | |||
| BacPhEn3 | 0.573 ± 0.084 | 4.51 × 104 | 2.37 × 104 | 8.73 × 104 | 2.95 | 1.03 | 11.40 | |
| OBs purified from larvae following injection with BVs | ||||||||
| BacPh | 0.555 ± 0.088 | 1.15 × 105 | 5.93 × 104 | 2.58 × 105 | 1.16 | 0.27 | 5.03 | |
| BacPhEn3 | 0.488 ± 0.093 | 2.03 × 105 | 8.73 × 104 | 7.52 × 105 | 0.65 | 0.13 | 3.33 | |
| Fourth instar | OBs and cell lysate from Sf9 cells | |||||||
| BacPh | 0.847 ± 0.126 | 1.10 × 106 | 6.15 × 105 | 2.84 × 106 | 1 | - | - | |
| BacPhEn3 | 1.019 ± 0.119 | 2.34 × 105 | 1.66 × 105 | 3.53 × 105 | 4.69 | 2.63 | 10.59 | |
| OBs purified from larvae following injection with BVs | ||||||||
| BacPh | 0.830 ± 0.110 | 1.09 × 106 | 6.38 × 105 | 2.14 × 106 | 1.01 | 0.40 | 2.07 | |
| BacPhEn3 | 0.756 ± 0.106 | 1.13 × 106 | 6.32 × 105 | 2.37 × 106 | 0.97 | 0.37 | 2.02 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricarte-Bermejo, A.; Simón, O.; Fernández, A.B.; Williams, T.; Caballero, P. Bacmid Expression of Granulovirus Enhancin En3 Accumulates in Cell Soluble Fraction to Potentiate Nucleopolyhedrovirus Infection. Viruses 2021, 13, 1233. https://doi.org/10.3390/v13071233
Ricarte-Bermejo A, Simón O, Fernández AB, Williams T, Caballero P. Bacmid Expression of Granulovirus Enhancin En3 Accumulates in Cell Soluble Fraction to Potentiate Nucleopolyhedrovirus Infection. Viruses. 2021; 13(7):1233. https://doi.org/10.3390/v13071233
Chicago/Turabian StyleRicarte-Bermejo, Adriana, Oihane Simón, Ana Beatriz Fernández, Trevor Williams, and Primitivo Caballero. 2021. "Bacmid Expression of Granulovirus Enhancin En3 Accumulates in Cell Soluble Fraction to Potentiate Nucleopolyhedrovirus Infection" Viruses 13, no. 7: 1233. https://doi.org/10.3390/v13071233
APA StyleRicarte-Bermejo, A., Simón, O., Fernández, A. B., Williams, T., & Caballero, P. (2021). Bacmid Expression of Granulovirus Enhancin En3 Accumulates in Cell Soluble Fraction to Potentiate Nucleopolyhedrovirus Infection. Viruses, 13(7), 1233. https://doi.org/10.3390/v13071233