Envelope-Fusion-Syncytium Formation in Microplitis bicoloratus bracovirus Maturation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Insect Ovaries
2.2. Histological Staining
2.3. Sample Preparation for TEM Analysis
2.4. Ultramicrotomy and Electron Microscopy
2.5. Cross-Sectional Area of Calyx Cell Nucleus and Fused Virus Envelopes
2.6. Statistical Analysis
3. Results
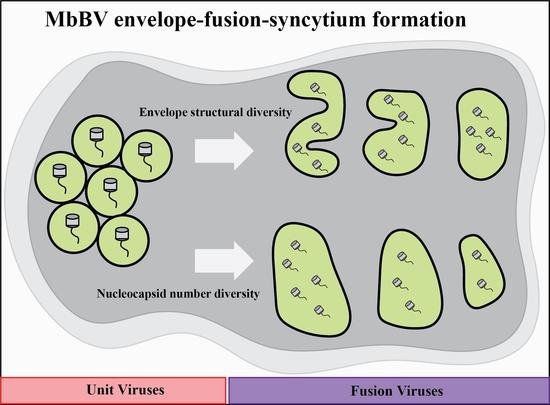
3.1. Microplitis Bicoloratus Bracovirus Envelope Synthesis and Fusion Process
3.2. Form of the Envelope-Fusion-Syncytium
3.3. PCECs Clear Non-Enveloped Nucleocapsids
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Strand, M.R.; Burke, G.R. Polydnavirus-wasp associations: Evolution, genome organization, and function. Curr. Opin. Virol. 2013, 3, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Fath-Goodin, A.; Webb, B.A. Polydnaviruses: General Features. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, UK, 2008; pp. 256–261. [Google Scholar] [CrossRef]
- Volkoff, A.-N.; Huguet, E. Polydnaviruses (Polydnaviridae); Academic Press: Cambridge, MA, USA, 2021; Volume 4, pp. 849–857. [Google Scholar]
- Volkoff, A.-N.; Drezen, J.-M.; Cusson, M.; Webb, B.A. Parasitoid Viruses: Symbiont and Pathogens; Academic Press: Cambridge, MA, USA, 2012; pp. 33–45. [Google Scholar]
- Wyler, T.; Lanzrein, B. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. II. Ultrastructural analysis of calyx cell development, virion formation and release. J. Gen. Virol. 2003, 84, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.J.; Frascaroli, G.; Zhou, X.; Knickmann, J.; Brune, W. Cell Fusion and Syncytium Formation in Betaherpesvirus Infection. Viruses 2021, 13, 1973. [Google Scholar] [CrossRef] [PubMed]
- Assi, W.; Hirose, T.; Wada, S.; Matsuura, R.; Takeshima, S.-N.; Aida, Y. PRMT5 Is Required for Bovine Leukemia Virus Infection In Vivo and Regulates BLV Gene Expression, Syncytium Formation, and Glycosylation In Vitro. Viruses 2020, 12, 650. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murria, M.J.; Exposito-Dominguez, N.; Duart, G.; Mingarro, I.; Martinez-Gil, L. A Bimolecular Multicellular Complementation System for the Detection of Syncytium Formation: A New Methodology for the Identification of Nipah Virus Entry Inhibitors. Viruses 2019, 11, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerna, G.; Percivalle, E.; Perez, L.; Lanzavecchia, A.; Lilleri, D. Monoclonal Antibodies to Different Components of the Human Cytomegalovirus (HCMV) Pentamer gH/gL/pUL128L and Trimer gH/gL/gO as well as Antibodies Elicited during Primary HCMV Infection Prevent Epithelial Cell Syncytium Formation. J. Virol. 2016, 90, 6216–6223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Huang, K.; Krishnan, S.; Svabek, C.; Rowe, D.C.; Brewah, Y.; Sanjuan, M.; Patera, A.C.; Kolbeck, R.; Herbst, R.; et al. RAGE inhibits human respiratory syncytial virus syncytium formation by interfering with F-protein function. J. Gen. Virol. 2013, 94, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Marti, D.; Grossniklaus-Bürgin, C.; Wyder, S.; Wyler, T.; Lanzrein, B. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. I. Ovary morphogenesis, amplification of viral DNA and ecdysteroid titres. J. Gen. Virol. 2003, 84, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.-H.; Chen, Y.-B.; Li, M.; Cai, Q.-C.; Zhang, L.-D.; Lu, Z.-Y.; Li, J.-C.; Zhu, Q.-S.; Ji, G.; Luo, K.-J. Cryo-EM structure reveals cylindrical nucleocapsids from two polydnaviruses. Arch. Virol. 2018, 163, 3357–3363. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.-J.; Pang, Y. Disruption Effect of Microplitis bicoloratus Polydnavirus EGF-like Protein, MbCRP, on Actin Cytoskeleton in Lepidopteran Insect Hemocytes. Acta Biochim. Biophys. Sin. 2006, 38, 577–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Buron, I.; Beckage, N.E. Characterization of a polydnavirus (PDV) and virus-like filamentous particle (VLFP) in the braconid wasp Cotesia congregata (Hymenoptera: Braconidae). J. Invertebr. Pathol. 1992, 59, 315–327. [Google Scholar] [CrossRef]
- Albrecht, W.T.; Pfister-Wilhelm, R.; Gruber, A.; Stettler, P.; Heiniger, P.; Kurt, E.; Schumperli, D.; Lanzrein, B. Polydnavirus of the parasitic wasp Chelonus inanitus (Braconidae): Characterization, genome organization and time point of replication. J. Gen. Virol. 1994, 75, 3353–3363. [Google Scholar] [CrossRef] [PubMed]
- Arvin, M.J.; Lorenzi, A.; Burke, G.R.; Strand, M.R. MdBVe46 is an envelope protein that is required for virion formation by Microplitis demolitor bracovirus. J. Gen. Virol. 2021, 102, 001565. [Google Scholar] [CrossRef] [PubMed]
- Blissard, G.W.; Wenz, J.R. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 1992, 66, 6829–6835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernomordik, L.; Leikina, E.; Cho, M.S.; Zimmerberg, J. Control of baculovirus gp64-induced syncytium formation by membrane lipid composition. J. Virol. 1995, 69, 3049–3058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Leikina, E.; Melikov, K.; Podbilewicz, B.; Kozlov, M.M.; Chernomordik, L.V. Fusion-pore expansion during syncytium formation is restricted by an actin network. J. Cell Sci. 2008, 121, 3619–3628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, B.A.; Strand, M.R.; Dickey, S.E.; Beck, M.H.; Hilgarth, R.S.; Barney, W.E.; Kadash, K.; Kroemer, J.A.; Lindstrom, K.G.; Rattanadechakul, W.; et al. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 2005, 347, 160–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espagne, E.; Dupuy, C.; Huguet, E.; Cattolico, L.; Provost, B.; Martins, N.; Poirié, M.; Periquet, G.; Drezen, J.M. Genome Sequence of a Polydnavirus: Insights into Symbiotic Virus Evolution. Science 2004, 306, 286–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, M.H.; Inman, R.B.; Strand, M.R. Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology 2007, 359, 179–189. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, M.-W.; Luo, K.-J. Envelope-Fusion-Syncytium Formation in Microplitis bicoloratus bracovirus Maturation. Viruses 2022, 14, 2183. https://doi.org/10.3390/v14102183
Dai M-W, Luo K-J. Envelope-Fusion-Syncytium Formation in Microplitis bicoloratus bracovirus Maturation. Viruses. 2022; 14(10):2183. https://doi.org/10.3390/v14102183
Chicago/Turabian StyleDai, Ming-Wu, and Kai-Jun Luo. 2022. "Envelope-Fusion-Syncytium Formation in Microplitis bicoloratus bracovirus Maturation" Viruses 14, no. 10: 2183. https://doi.org/10.3390/v14102183
APA StyleDai, M. -W., & Luo, K. -J. (2022). Envelope-Fusion-Syncytium Formation in Microplitis bicoloratus bracovirus Maturation. Viruses, 14(10), 2183. https://doi.org/10.3390/v14102183