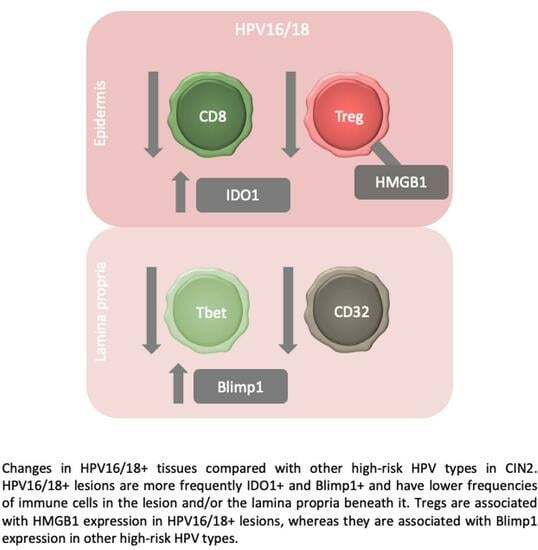
The High-Risk Human Papillomavirus Type Influences the Tissue Microenvironment in Cervical Intraepithelial Neoplasia Grade 2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. HPV Testing
2.3. Immunohistochemical Staining
2.4. Statistical Analysis
3. Results
3.1. HPV16-Positive Lesions Were Frequently Also Positive for ‘Other’ hr HPV
3.2. HPV16/18-Positive Women Are More Likely to Progress to More Severe Disease than HPV ‘Other’-Positive Women
3.3. E1^E4 Staining Was Infrequent in hrHPV-Positive CIN2 Lesions
3.4. IDO1 Expression Is More Frequent in HPV16/18-Positive Lesions
3.5. Immune Cell Infiltrates Were Reduced in HPV16/18 Lesions Compared with HPV ‘Other’ Lesions
3.6. The Correlations between Cells Differ in HPV16/18 and HPV ‘Other’ Tissues
3.7. The Lamina Propria Beneath IDO1-Positive Tissue Has Fewer CD4, CD8 T Cells and FoxP3-Positive Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iarc Working Group on the Evaluation of Carcinogenic Risks to Humans. IARC monographs on the evaluation of carcinogenic risks to humans. Biological Agents. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100B, 255–314. [Google Scholar]
- Karube, A.; Saito, F.; Waga, M.; Yokoyama, S.; Kanamori, K. Progression of cervical intraepithelial neoplasia grade 2 lesions among Japanese women harboring different genotype categories of high-risk human papillomaviruses. J. Rural. Med. 2021, 16, 91–97. [Google Scholar] [CrossRef]
- Kjaer, S.K.; Frederiksen, K.; Munk, C.; Iftner, T. Long-term absolute risk of cervical intraepithelial neoplasia grade 3 or worse following human papillomavirus infection: Role of persistence. J. Natl. Cancer Inst. 2010, 102, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. 1), 2–23. [Google Scholar] [CrossRef]
- Matthews, K.; Leong, C.M.; Baxter, L.; Inglis, E.; Yun, K.; Backstrom, B.T.; Doorbar, J.; Hibma, M. Depletion of Langerhans cells in human papillomavirus type 16-infected skin is associated with E6-mediated down regulation of E-cadherin. J. Virol. 2003, 77, 8378–8385. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, E.M.; Lee, E.H.; Ji, K.Y.; Yi, J.; Park, M.; Kim, K.D.; Cho, Y.Y.; Kang, H.S. Human papillomavirus 16E6 suppresses major histocompatibility complex class I by upregulating lymphotoxin expression in human cervical cancer cells. Biochem. Biophys. Res. Commun. 2011, 409, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.L.; Woodby, B.L.; Ulicny, J.; Raikhy, G.; Orr, A.W.; Songock, W.K.; Bodily, J.M. Human Papillomavirus 16 E5 Inhibits Interferon Signaling and Supports Episomal Viral Maintenance. J. Virol. 2020, 94, e01582-19. [Google Scholar] [CrossRef] [PubMed]
- Bortnik, V.; Wu, M.; Julcher, B.; Salinas, A.; Nikolic, I.; Simpson, K.J.; McMillan, N.A.; Idris, A. Loss of HPV type 16 E7 restores cGAS-STING responses in human papilloma virus-positive oropharyngeal squamous cell carcinomas cells. J. Microbiol. Immunol. Infect. 2021, 54, 733–739. [Google Scholar] [CrossRef]
- Miyauchi, S.; Kim, S.S.; Jones, R.N.; Zhang, L.; Guram, K.; Sharma, S.; Schoenberger, S.P.; Cohen, E.E.W.; Califano, J.A.; Sharabi, A.B. Human papillomavirus E5 suppresses immunity via inhibition of the immunoproteasome and STING pathway. Cell Rep. 2023, 42, 112508. [Google Scholar] [CrossRef]
- Mittal, D.; Kassianos, A.J.; Tran, L.S.; Bergot, A.S.; Gosmann, C.; Hofmann, J.; Blumenthal, A.; Leggatt, G.R.; Frazer, I.H. Indoleamine 2,3-dioxygenase activity contributes to local immune suppression in the skin expressing human papillomavirus oncoprotein e7. J. Investig. Dermatol. 2013, 133, 2686–2694. [Google Scholar] [CrossRef]
- Munn, D.H.; Shafizadeh, E.; Attwood, J.T.; Bondarev, I.; Pashine, A.; Mellor, A.L. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 1999, 189, 1363–1372. [Google Scholar] [CrossRef]
- Munn, D.H.; Zhou, M.; Attwood, J.T.; Bondarev, I.; Conway, S.J.; Marshall, B.; Brown, C.; Mellor, A.L. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 1998, 281, 1191–1193. [Google Scholar] [CrossRef]
- Schafer, C.C.; Wang, Y.; Hough, K.P.; Sawant, A.; Grant, S.C.; Thannickal, V.J.; Zmijewski, J.; Ponnazhagan, S.; Deshane, J.S. Indoleamine 2,3-dioxygenase regulates anti-tumor immunity in lung cancer by metabolic reprogramming of immune cells in the tumor microenvironment. Oncotarget 2016, 7, 75407–75424. [Google Scholar] [CrossRef]
- Liu, H.; Liu, L.; Liu, K.; Bizargity, P.; Hancock, W.W.; Visner, G.A. Reduced cytotoxic function of effector CD8+ T cells is responsible for indoleamine 2,3-dioxygenase-dependent immune suppression. J. Immunol. 2009, 183, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Rajesh, A.; Innes, C.; van der Griend, R.; Fitzgerald, P.; Simcock, B.; Sykes, P.; Hibma, M. Blimp-1 is a prognostic indicator for progression of cervical intraepithelial neoplasia grade 2. J. Cancer Res. Clin. Oncol. 2022, 148, 1991–2002. [Google Scholar] [CrossRef] [PubMed]
- Sykes, P.; Innes, C.; Harker, D.; Whitehead, M.; van der Griend, R.; Lawton, B.; Hibma, M.; Fitzgerald, P.; Dudley, N.; Petrich, S.; et al. Observational Management of CIN 2 in Young Women: A Prospective Multicenter Trial. J. Low. Genit. Tract. Dis. 2016, 20, 343–347. [Google Scholar] [CrossRef]
- Innes, C.R.; Sykes, P.H.; Harker, D.; Williman, J.A.; Van der Griend, R.A.; Whitehead, M.; Hibma, M.; Lawton, B.A.; Fitzgerald, P.; Dudley, N.M.; et al. Changes in human papillomavirus genotypes associated with cervical intraepithelial neoplasia grade 2 lesions in a cohort of young women (2013–2016). Papillomavirus Res. 2018, 6, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Davy, C.E.; Jackson, D.J.; Raj, K.; Peh, W.L.; Southern, S.A.; Das, P.; Sorathia, R.; Laskey, P.; Middleton, K.; Nakahara, T.; et al. Human papillomavirus type 16 E1 E4-induced G2 arrest is associated with cytoplasmic retention of active Cdk1/cyclin B1 complexes. J. Virol. 2005, 79, 3998–4011. [Google Scholar] [CrossRef]
- Raj, K.; Berguerand, S.; Southern, S.; Doorbar, J.; Beard, P. E1 empty set E4 protein of human papillomavirus type 16 associates with mitochondria. J. Virol. 2004, 78, 7199–7207. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Foo, C.; Coleman, N.; Medcalf, L.; Hartley, O.; Prospero, T.; Napthine, S.; Sterling, J.; Winter, G.; Griffin, H. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 1997, 238, 40–52. [Google Scholar] [CrossRef]
- Keating, J.T.; Cviko, A.; Riethdorf, S.; Riethdorf, L.; Quade, B.J.; Sun, D.; Duensing, S.; Sheets, E.E.; Munger, K.; Crum, C.P. Ki-67, cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am. J. Surg. Pathol. 2001, 25, 884–891. [Google Scholar] [CrossRef]
- Tang, K.; Wu, Y.H.; Song, Y.; Yu, B. Indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors in clinical trials for cancer immunotherapy. J. Hematol. Oncol. 2021, 14, 68. [Google Scholar] [CrossRef]
- Akinleye, A.; Rasool, Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Tainio, K.; Athanasiou, A.; Tikkinen, K.A.O.; Aaltonen, R.; Hernandes, J.C.; Glazer-Livson, S.; Jakobsson, M.; Joronen, K.; Kiviharju, M.; Louvanto, K.; et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: Systematic review and meta-analysis. BMJ 2018, 360, k499. [Google Scholar] [CrossRef] [PubMed]
- Sykes, P.H.; Simcock, B.J.; Innes, C.R.; Harker, D.; Williman, J.A.; Whitehead, M.; van der Griend, R.A.; Lawton, B.A.; Hibma, M.; Fitzgerald, P.; et al. Predicting regression of cervical intraepithelial neoplasia grade 2 in women under 25 years. Am. J. Obstet. Gynecol. 2022, 226, 222.e1–222.e13. [Google Scholar] [CrossRef]
- Darragh, T.M.; Colgan, T.J.; Thomas Cox, J.; Heller, D.S.; Henry, M.R.; Luff, R.D.; McCalmont, T.; Nayar, R.; Palefsky, J.M.; Stoler, M.H.; et al. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: Background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Int. J. Gynecol. Pathol. 2013, 32, 76–115. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Drubin, M.E.; Crum, C.P.; Munger, K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. USA 2011, 108, 2130–2135. [Google Scholar] [CrossRef]
- Doorbar, J. Host control of human papillomavirus infection and disease. Best. Pract. Res. Clin. Obstet. Gynaecol. 2018, 47, 27–41. [Google Scholar] [CrossRef]
- Theate, I.; van Baren, N.; Pilotte, L.; Moulin, P.; Larrieu, P.; Renauld, J.C.; Herve, C.; Gutierrez-Roelens, I.; Marbaix, E.; Sempoux, C.; et al. Extensive profiling of the expression of the indoleamine 2,3-dioxygenase 1 protein in normal and tumoral human tissues. Cancer Immunol. Res. 2015, 3, 161–172. [Google Scholar] [CrossRef]
- Heeren, A.M.; van Dijk, I.; Berry, D.; Khelil, M.; Ferns, D.; Kole, J.; Musters, R.J.P.; Thijssen, V.L.; Mom, C.H.; Kenter, G.G.; et al. Indoleamine 2,3-Dioxygenase Expression Pattern in the Tumor Microenvironment Predicts Clinical Outcome in Early Stage Cervical Cancer. Front. Immunol. 2018, 9, 1598. [Google Scholar] [CrossRef]
- Venancio, P.A.; Consolaro, M.E.L.; Derchain, S.F.; Boccardo, E.; Villa, L.L.; Maria-Engler, S.S.; Campa, A.; Discacciati, M.G. Indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase expression in HPV infection, SILs, and cervical cancer. Cancer Cytopathol. 2019, 127, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Succaria, F.; Kvistborg, P.; Stein, J.E.; Engle, E.L.; McMiller, T.L.; Rooper, L.M.; Thompson, E.; Berger, A.E.; van den Brekel, M.; Zuur, C.L.; et al. Characterization of the tumor immune microenvironment in human papillomavirus-positive and -negative head and neck squamous cell carcinomas. Cancer Immunol. Immunother. 2021, 70, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Rader, J.S.; Zhang, F.; Liapis, H.; Koki, A.T.; Masferrer, J.L.; Subbaramaiah, K.; Dannenberg, A.J. Cyclooxygenase-2 is overexpressed in human cervical cancer. Clin. Cancer Res. 2001, 7, 429–434. [Google Scholar] [PubMed]
- Muntinga, C.L.P.; de Vos van Steenwijk, P.J.; Bekkers, R.L.M.; van Esch, E.M.G. Importance of the Immune Microenvironment in the Spontaneous Regression of Cervical Squamous Intraepithelial Lesions (cSIL) and Implications for Immunotherapy. J. Clin. Med. 2022, 11, 1432. [Google Scholar] [CrossRef]
- Malviya, V.; Yshii, L.; Junius, S.; Garg, A.D.; Humblet-Baron, S.; Schlenner, S.M. Regulatory T-cell stability and functional plasticity in health and disease. Immunol. Cell Biol. 2023, 101, 112–129. [Google Scholar] [CrossRef] [PubMed]



| HPV16/18 (n = 30) | HPV ‘Other’ (n = 39) | Statistical Significance | ||
|---|---|---|---|---|
| Age | Mean (Range) | 21.50 (17–24) | 21.91 (19–24) | p = 0.1043 (M–W U *) |
| Smoker | Yes | 13 (43.3%) | 11 (28.2%) | p = 0.4031 (chi-square) |
| No | 17 (56.7%) | 26 (66.7%) | ||
| Unknown | - | 2 (5.1%) | ||
| Vaccine | Yes | 2 (6.7%) | 25 (64.1%) | p < 0.0001 (chi-square) |
| No | 12 (40.0%) | 4 (10.3%) | ||
| Unknown | 16 (53.3%) | 10 (25.6%) | ||
| Lesion area | Mean (Range) | 0.4334 (0.0093–2.148) | 0.3812 (0.0112–1.672) | p = 0.9462 (M–W U *) |
| HPV16/18 (n = 30) | HPV ‘Other’ (n = 39) | ||||||
|---|---|---|---|---|---|---|---|
| Ki67 | E4 | Ki67 | E4 | ||||
| 1/3 * | 5 (16.7%) | E4+ | 0 (0%) | 1/3 | 8 (21.1%) | E4+ | 4 (57.1%) |
| E4− | 5 (100%) | E4− | 3 (42.9%) | ||||
| ND ** | 1 | ND | 1 | ||||
| 2/3 | 23 (76.7%) | E4+ | 3 (13.0%) | 2/3 | 24 (63.2%) | E4+ | 4 (17.4%) |
| E4− | 20 (87.0%) | E4− | 19 (82.6%) | ||||
| ND | 1 | ND | 1 | ||||
| 3/3 | 2 (6.7%) | E4+ | 0 (0%) | 3/3 | 6 (15.8%) | E4+ | 0 (0%) |
| E4− | 2 (100%) | E4− | 6 (100%) | ||||
| ND | - | ND | - | ||||
| ND | ND | 1 | E4− | 1 | |||
| HPV16/18 (n = 30) | HPV ‘Other’ (n = 39) | Fisher’s Exact Test | ||||||
|---|---|---|---|---|---|---|---|---|
| PD-L1 | ||||||||
| Positive | 5 (16.7%) | * <CIN3 | 1 (20.0%) | 7 (18.4%) | <CIN3 | 5 (71.4%) | p = 0.8505 | |
| CIN3 | 4 (80.0%) | CIN3 | 2 (28.6%) | |||||
| Negative | 25 (83.3%) | <CIN3 | 7 (28.0%) | 31 (81.6%) | <CIN3 | 21 (67.7%) | ||
| CIN3 | 18 (72.0%) | CIN3 | 10 (32.3%) | |||||
| IDO1 | ||||||||
| Positive | 14 (46.7%) | <CIN3 | 4 (28.6%) | 2 (5.4%) | <CIN3 | 1 (50.0%) | p < 0.0001 | |
| CIN3 | 10 (71.4%) | CIN3 | 1 (50.0%) | |||||
| Negative | 16 (53.3%) | <CIN3 | 4 (25.0%) | 35 (94.6%) | <CIN3 | 24 (68.6%) | ||
| CIN3 | 12 (75.0%) | CIN3 | 11 (31.4%) | |||||
| Epidermis | Lamina Propria | |||||
|---|---|---|---|---|---|---|
| Cells/mm2 (Mean ± SD) | Cells/mm2 (Mean ± SD) | |||||
| HPV16/18 | HPV ‘Other’ | p Value # | HPV16/18 | HPV ‘Other’ | p Value # | |
| CD4 | 298.13 ± 237.48 | 407.11 ± 380.94 | 0.2072 | 2008.57 ± 1986.88 | 1993.33 ± 1506.65 | 0.7521 |
| Tbet | 88.74 ± 117.54 | 151.09 ± 146.81 | 0.1013 | 371.78 ± 276.78 | 834.04 ± 945.68 | 0.0335 * |
| GATA3 | 1579.15 ± 1968.70 | 1223.91 ± 1346.17 | 0.7189 | 3009.15 ± 2279.40 | 2399.54 ± 1665.51 | 0.3533 |
| IL17 | 489.24 ± 664.42 | 380.62 ± 481.35 | 0.6273 | 1280.58 ± 680.08 | 1613.62 ± 851.25 | 0.1329 |
| FoxP3 | 27.23 ± 100.09 | 24.61 ± 45.77 | 0.4164 | 56.27 ± 69.83 | 79.59 ± 79.14 | 0.2534 |
| FoxP3+CD4− | 21.35 ± 99.16 | 12.49 ± 39.86 | 0.8742 | 17.21 ± 28.94 | 17.50 ± 24.48 | 0.6946 |
| FoxP3+CD4+ | 5.15 ± 9.15 | 12.12 ± 14.54 | 0.0151 * | 39.06 ± 50.12 | 62.09 ± 65.94 | 0.1555 |
| CD8 | 125.09 ± 104.54 | 246.16 ± 210.55 | 0.0086 ** | 609.07 ± 454.41 | 888.83 ± 804.86 | 0.1275 |
| Granzyme B | 95.85 ± 127.42 | 129.85 ± 144.74 | 0.2477 | 411.28 ± 417.18 | 605.03 ± 817.21 | 0.3997 |
| CD8+ GranzymeB+ | 0.54 ± 1.27 | 1.89 ± 4.39 | 0.1944 | 11.83 ± 23.91 | 66.75 ± 227.14 | 0.1811 |
| Langerin+ | 21.70 ± 26.78 | 38.41 ± 58.94 | 0.194 | 2.92 ± 5.54 | 6.05 ± 9.65 | 0.2664 |
| Langerin+ Fascin+ | 12.48 ± 14.76 | 26.91 ± 43.32 | 0.193 | 1.10 ± 2.97 | 2.36 ± 5.65 | 0.2948 |
| CD11c | 87.07 ± 66.39 | 109.04 ± 126.70 | 0.8521 | 545.86 ± 1621.87 | 353.28 ± 341.68 | 0.9857 |
| CD32 | 195.57 ± 473.50 | 408.62 ± 866.34 | 0.0644 | 80.04 ± 74.57 | 235.32 ± 297.03 | 0.0052 ** |
| CD138 | 740.95 ± 642.07 | 1043.74 ± 1912.31 | 0.4895 | 845.84 ± 715.60 | 1706.27 ± 2549.27 | 0.1554 |
| HMGB1 | 1615.30 ± 1559.87 | 1534.52 ± 1364.22 | 0.7463 | 3790.87 ± 1989.52 | 3841.23 ± 1995.40 | 0.9285 |
| Blimp1 | 537.32 ± 996.82 | 343.93 ± 1094.08 | 0.1097 | 1760.00 ± 1563.05 | 1033.87 ± 1392.86 | 0.0447 * |
| TSLP | a ND | ND | ND | 1205.08 ± 793.24 | 941.04 ± 647.56 | 0.207 |
| Area | 0.43 ± 0.49 | 0.38 ± 0.42 | 0.748 | ND | ND | ND |
| Epidermis | Lamina Propria | |||||
|---|---|---|---|---|---|---|
| IDO+ Cells/mm2 | IDO− Cells/mm2 | p Value # | IDO+ Cells/mm2 | IDO− Cells/mm2 | p Value # | |
| CD4 | 304.57 | 377.51 | 0.408 | 1075.66 | 2265.73 | 0.01 ** |
| Tbet | 92.61 | 134.41 | 0.6268 | 344.55 | 740.23 | 0.1176 |
| GATA3 | 1384.55 | 1405.66 | 0.9136 | 2999.24 | 2546.18 | 0.4573 |
| IL17 | 426.39 | 425.44 | 0.7062 | 1744.54 | 1377.64 | 0.1172 |
| FoxP3 | 10.55 | 30.84 | 0.5006 | 21.80 | 85.69 | 0.0006 *** |
| FoxP3+CD4− | 3.53 | 21.10 | 0.5685 | 3.95 | 22.40 | 0.0034 ** |
| FoxP3+CD4+ | 6.79 | 9.74 | 0.5781 | 17.85 | 63.29 | 0.0023 ** |
| CD8 | 163.56 | 201.05 | 0.6991 | 477.46 | 874.07 | 0.0304 * |
| Granzyme B | 93.63 | 117.03 | 0.6063 | 357.19 | 585.70 | 0.4148 |
| CD8+ GranzymeB+ | 0.88 | 1.45 | 0.7508 | 10.73 | 58.24 | 0.1161 |
| Langerin+ | 22.00 | 34.30 | 0.3968 | 2.50 | 5.37 | 0.4949 |
| Langerin+ Fascin+ | 14.42 | 22.83 | 0.6646 | 1.29 | 1.83 | 0.6469 |
| CD11c | 83.50 | 105.68 | 0.6951 | 193.95 | 506.50 | 0.084 |
| CD32 | 195.57 | 367.30 | 0.4119 | 91.10 | 197.40 | 0.1652 |
| CD138 | 616.64 | 1014.23 | 0.3613 | 691.26 | 1549.87 | 0.1375 |
| HMGB1 | 1823.29 | 1448.84 | 0.1634 | 3796.03 | 3778.15 | 0.7355 |
| Blimp1 | 943.97 | 281.20 | 0.0796 | 1634.13 | 1236.52 | 0.2363 |
| TSLP | a ND | ND | ND | 1105.43 | 1028.97 | 0.7995 |
| Area | 0.41 | 0.40 | 0.5534 | ND | ND | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, M.; Rajesh, A.; Innes, C.; van der Griend, R.; Fitzgerald, P.; Simcock, B.; Sykes, P.; Hibma, M. The High-Risk Human Papillomavirus Type Influences the Tissue Microenvironment in Cervical Intraepithelial Neoplasia Grade 2. Viruses 2023, 15, 1953. https://doi.org/10.3390/v15091953
Saito M, Rajesh A, Innes C, van der Griend R, Fitzgerald P, Simcock B, Sykes P, Hibma M. The High-Risk Human Papillomavirus Type Influences the Tissue Microenvironment in Cervical Intraepithelial Neoplasia Grade 2. Viruses. 2023; 15(9):1953. https://doi.org/10.3390/v15091953
Chicago/Turabian StyleSaito, Mayumi, Aarthi Rajesh, Carrie Innes, Rachael van der Griend, Peter Fitzgerald, Bryony Simcock, Peter Sykes, and Merilyn Hibma. 2023. "The High-Risk Human Papillomavirus Type Influences the Tissue Microenvironment in Cervical Intraepithelial Neoplasia Grade 2" Viruses 15, no. 9: 1953. https://doi.org/10.3390/v15091953
APA StyleSaito, M., Rajesh, A., Innes, C., van der Griend, R., Fitzgerald, P., Simcock, B., Sykes, P., & Hibma, M. (2023). The High-Risk Human Papillomavirus Type Influences the Tissue Microenvironment in Cervical Intraepithelial Neoplasia Grade 2. Viruses, 15(9), 1953. https://doi.org/10.3390/v15091953