A New Validation Methodology for In Silico Tools Based on X-ray Computed Tomography Images of Tablets and a Performance Analysis of One Tool
Abstract
:1. Introduction
2. Materials and Methods
2.1. Excipients and Tableting
2.2. Characterisation
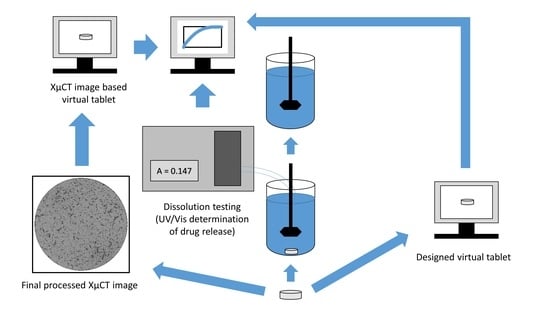
2.3. Dissolution Testing
2.4. Imaging and Image Processing
Nomenclature and Methods of Pathways
2.5. Matrices of the In Silico Dissolution Calculation
2.6. Parameters of the In Silico Dissolution Calculation
2.7. Analysis of the Dissolution Profiles
2.7.1. Difference and Similarity Factors
2.7.2. Kinetic Analysis
3. Results and Discussion
3.1. Determined Parameters
3.2. Computation Performance
3.3. Batch T25E75
3.4. Batch T50E50
3.5. Batch T75E25
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Tables
| Name | Tablet | Pathway | Tablet Desirability | API | Excipient |
|---|---|---|---|---|---|
| mg | mg | ||||
| real | T50E50-01 | - | - | 203.8 | 229.7 |
| B1 | T50E50-01 | Ref/UC/NLM/EC2/G2.3/H-iW//O-iB | 0.995 | 201.4 | 235.3 |
| B2 | T50E50-01 | Ref/PFF/UC/G2.2/EC11/KM(11) | 1.000 | 207.2 | 229.9 |
| D | T50E50-01 | - | - | 88.6 | 316.2 |
| G | T50E50-01 | - | - | 88.6 | 316.2 |
| real | T50E50-03 | - | - | 199.9 | 212.4 |
| B1 | T50E50-03 | Ref/UC/NLM/EC2/G2.3/H-iW//O-iB | 1.000 | 196.9 | 215.9 |
| B2 | T50E50-03 | Ref/PFF/UC/G2.2/EC11/KM(11) | 0.990 | 194.3 | 209.5 |
| B3 | T50E50-03 | Ref/PFF/UC/G3.7/EC16/H-iW//Y-iBiW | 0.978 | 194.2 | 217.4 |
| B4 | T50E50-03 | Ref/UC/NLM/G3.0/EC16/ID-iB//O-iB | 0.989 | 195.7 | 217.8 |
| C1 | T50E50-03 | Sta/RB/UC/EC17/G1.5/KM(12) | 0.747 | 218.3 | 198.9 |
| C2 | T50E50-03 | Sta/UC/UC/EC13/G2.5/KM(12) | 0.683 | 175.0 | 202.4 |
| API1 | T50E50-03 | Ref/PFF/UC/G3.3/EC13/KM(12) | 0.987 | 205.2 | 207.9 |
| API2 | T50E50-03 | Ref/PFF/UC/EC16/G3.7/KM(12) | 0.962 | 206.7 | 206.3 |
| T1 | T50E50-03 | Ref/UC/NLM/EC15/G2.9/H-iBiW//ID-iB | 1.000 | 200.0 | 212.4 |
| T2 | T50E50-03 | Sta/PFF/NLM/EC15/G2.8/KM(11) | 1.000 | 200.9 | 213.4 |
| T3 | T50E50-03 | Ref/PFF/NLM/EC18/G1.6/O-iB//Y-iBiW | 0.900 | 209.5 | 202.7 |
| T4 | T50E50-03 | Ref/PFF/UC/G3.5/EC19/KM(5) | 0.800 | 213.1 | 197.7 |
| T5 | T50E50-03 | Sta/RB/UC/G1.7/EC18/MaE-iW//MiE-iB | 0.700 | 181.6 | 228.2 |
| T6 | T50E50-03 | Ref/PFF/NLM/EC17/G3/MaE-iB//MiE-iB | 0.600 | 219.1 | 188.4 |
| T7 | T50E50-03 | Ref/UC/NLM/G3.1/EC18/MaE-iBiW//MiE-iB | 0.500 | 218.8 | 182.8 |
| T8 | T50E50-03 | Sta/RB/UC/G1.6/EC17/D-iB//MaE-iBiW | 0.400 | 230.0 | 187.4 |
| T9 | T50E50-03 | Ref/PFF/NLM/EC18/G3.1/MiE-iB//O | 0.300 | 218.9 | 177.8 |
| T10 | T50E50-03 | Ref/UC/NLM/EC16/G3.0/D//MaE | 0.201 | 232.0 | 179.7 |
| T11 | T50E50-03 | Ref/UC/NLM/G3.2/EC18/D//MaE | 0.101 | 232.8 | 178.2 |
| D | T50E50-03 | - | - | 88.6 | 316.2 |
| G | T50E50-03 | - | - | 88.6 | 316.2 |
| real | T50E50-05 | - | - | 201.2 | 218.3 |
| B1 | T50E50-05 | Ref/UC/NLM/EC2/G2.3/H-iW//O-iB | 1.000 | 204.3 | 217.7 |
| B2 | T50E50-05 | Ref/PFF/UC/G2.2/EC11/KM(11) | 1.000 | 199.8 | 220.6 |
| D | T50E50-05 | - | - | 88.6 | 316.2 |
| G | T50E50-05 | - | - | 88.6 | 316.2 |
| Name | Tablet | Pathway | Tablet Desirability | API | Excipient |
|---|---|---|---|---|---|
| mg | mg | ||||
| real | T75E25-01 | - | - | 319.6 | 109.1 |
| B1 | T75E25-01 | Ref/PFF/NLM/G1.5/EC16/KM(12) | 1.000 | 313.6 | 108.7 |
| B4 | T75E25-01 | Sta/RB/UC/EC18/G1.7/KM(9) | 0.986 | 314.1 | 112.2 |
| D | T75E25-01 | - | - | 88.6 | 316.2 |
| G | T75E25-01 | - | - | 88.6 | 316.2 |
| real | T75E25-03 | - | - | 318.3 | 115.9 |
| B1 | T75E25-03 | Ref/PFF/NLM/G1.5/EC16/KM(12) | 0.998 | 317.1 | 118.3 |
| B2 | T75E25-03 | Ref/PFF/NLM/G2.7/EC15/KM(12) | 1.000 | 320.5 | 116.9 |
| B3 | T75E25-03 | Ref/PFF/UC/G2.0/EC13/KM(12) | 1.000 | 320.5 | 117.4 |
| B4 | T75E25-03 | Sta/RB/UC/EC18/G1.7/KM(9) | 1.000 | 316.5 | 115.8 |
| C1 | T75E25-03 | Sta/RB/UC/EC17/G1.5/KM(12) | 0.928 | 308.4 | 121.0 |
| C2 | T75E25-03 | Sta/UC/UC/EC13/G2.5/KM(12) | 0.975 | 310.9 | 119.4 |
| API1 | T75E25-03 | Ref/PFF/UC/G3.3/EC13/KM(12) | 1.000 | 321.7 | 115.0 |
| API2 | T75E25-03 | Ref/PFF/UC/EC16/G3.7/KM(12) | 1.000 | 321.6 | 114.6 |
| T1 | T75E25-03 | Ref/RB/NLM/G3.0/EC17/KM(12) | 1.000 | 320.6 | 116.7 |
| T2 | T75E25-03 | Ref/UC/NLM/EC16/G3.0/H-iB//MiE-iB | 1.000 | 319.9 | 116.4 |
| T3 | T75E25-03 | Sta/UC/UC/G2.7/EC13/P//T | 0.900 | 316.7 | 124.2 |
| T4 | T75E25-03 | Sta/RB/UC/G3.2/EC17/H-iW//R-iBiW | 0.800 | 308.6 | 126.9 |
| T5 | T75E25-03 | Ref/PFF/NLM/G1.6/EC17/H-iBiW//M | 0.700 | 281.7 | 112.2 |
| T6 | T75E25-03 | Sta/PFF/UC/G3.0/EC17/D-iW//O-iBiW | 0.600 | 285.9 | 126.7 |
| T7 | T75E25-03 | Ref/PFF/NLM/EC18/G3.1/L-iB//P-iW | 0.500 | 345.3 | 99.5 |
| T8 | T75E25-03 | Sta/UC/UC/G3.0/EC17/L//Y-iB | 0.400 | 348.9 | 98.1 |
| T9 | T75E25-03 | Sta/UC/UC/EC16/G2.8/M-iBiW//M-iW | 0.301 | 275.4 | 97.1 |
| T10 | T75E25-03 | Sta/UC/UC/EC16/G2.8/M-iW//R-iW | 0.201 | 267.0 | 97.9 |
| T11 | T75E25-03 | Ref/PFF/NLM/EC18/G1.7/M-iW//P-iBiW | 0.102 | 266.0 | 133.7 |
| D | T75E25-03 | - | - | 88.6 | 316.2 |
| G | T75E25-03 | - | - | 88.6 | 316.2 |
| real | T75E25-05 | - | - | 86.1 | 331.4 |
| B1 | T75E25-05 | Ref/PFF/NLM/G1.5/EC16/KM(12) | 1.000 | 307.3 | 116.3 |
| B4 | T75E25-05 | Sta/RB/UC/EC18/G1.7/KM(9) | 1.000 | 311.1 | 115.1 |
| D | T75E25-05 | - | - | 88.6 | 316.2 |
| G | T75E25-05 | - | - | 88.6 | 316.2 |
| -Value | -Value | Slope | Intercept | p-Value Slope | p-Value Intercept | ||
|---|---|---|---|---|---|---|---|
| - | - | 1.7517 0.4781 | 1.1117 0.3143 | 0.9993 0.9985 | - | - | |
| 10.6 | 69.6 | 2.5137 0.7463 | −17.0895 −0.4257 | 0.9964 0.9994 | 2.76 × 10 6.19 × 10 | NA NA | |
| 17.3 | 58.0 | 2.6400 0.6396 | −10.3343 −0.0398 | 0.9996 0.9991 | 3.98 × 10 2.05 × 10 | NA NA | |
| 33.3 | 42.9 | NA NA | NA NA | NA NA | NA NA | NA NA | |
| 122.5 | 19.5 | 6.8476 0.4314 | 8.0960 1.0233 | 0.9621 0.9703 | 1.55 × 10 8.99 × 10 | NA NA | |
| - | - | 2.2279 0.4709 | 2.3324 0.4465 | 0.9996 0.9997 | - | - | |
| 54.5 | 29.3 | 4.8015 0.7592 | −19.5489 −0.0649 | 0.9993 0.9995 | 2.77 × 10 1.12 × 10 | NA NA | |
| 55.3 | 29.4 | 4.7346 0.7169 | −16.0672 0.0573 | 0.9996 0.9993 | 5.57 × 10 8.84 × 10 | NA NA | |
| 125.6 | 13.1 | NA NA | NA NA | NA NA | NA NA | NA NA | |
| 128.1 | 12.8 | NA NA | NA NA | NA NA | NA NA | NA NA | |
| - | - | 5.6369 0.7242 | −16.7140 0.1437 | 0.9955 0.9992 | - | - | |
| 7.6 | 62.9 | 5.6819 0.7819 | −21.3730 −0.0204 | 0.9999 0.9983 | 6.73 × 10 4.86 × 10 | 3.65 × 10 NA | |
| 7.7 | 62.1 | 5.6879 0.7980 | −22.5207 −0.0669 | 0.9998 0.9985 | 6.18 × 10 3.19 × 10 | 1.33 × 10 NA | |
| 42.5 | 20.8 | NA NA | NA NA | NA NA | NA NA | NA NA | |
| 42.4 | 20.9 | NA NA | NA NA | NA NA | NA NA | NA NA |
| Matrix | Tablet Desirability | -Value | -Value | Slope | Intercept | p-Value Slope | p-Value Intercept | |
|---|---|---|---|---|---|---|---|---|
| real data | - | - | - | 2.1984 0.4677 | 2.4137 0.4501 | 0.9998 0.9996 | - | - |
| 0.987 | 59.9 | 28.0 | 4.9139 0.7326 | −17.3007 0.0331 | 0.9996 0.9993 | 3.33 × 10 4.62 × 10 | NA NA | |
| 0.962 | 60.3 | 27.9 | 4.9299 0.7324 | −17.3387 0.0351 | 0.9996 0.9993 | 9.79 × 10 1.01 × 10 | NA NA | |
| 0.747 | 61.3 | 27.1 | 5.0006 0.7760 | −20.7285 −0.0873 | 0.9993 0.9995 | 9.77 × 10 2.35 × 10 | NA NA | |
| 0.683 | 62.9 | 26.6 | 5.0293 0.7707 | −19.7255 −0.0614 | 0.9989 0.9996 | 1.19 × 10 1.72 × 10 | NA NA | |
| 1.000 | 57.6 | 28.3 | 4.9234 0.7770 | −20.7390 −0.1004 | 0.9990 0.9996 | 5.67 × 10 2.35 × 10 | NA NA | |
| 0.990 | 59.7 | 28.0 | 4.9072 0.7333 | −17.3619 0.0299 | 0.9995 0.9995 | 1.19 × 10 8.14 × 10 | NA NA | |
| 0.978 | 57.3 | 28.9 | 4.8021 0.7280 | −16.9167 0.0325 | 0.9995 0.9994 | 8.54 × 10 2.46 × 10 | NA NA | |
| 0.989 | 57.0 | 28.5 | 4.9549 0.7912 | −21.7382 −0.1390 | 0.9990 0.9995 | 4.46 × 10 5.69 × 10 | NA NA | |
| 1.000 | 58.7 | 28.0 | 4.9945 0.7849 | −21.3789 −0.1153 | 0.9991 0.9995 | 1.27 × 10 4.88 × 10 | NA NA | |
| 1.000 | 60.4 | 27.9 | 4.9825 0.7323 | −17.4912 0.0402 | 0.9996 0.9992 | 7.95 × 10 8.65 × 10 | NA NA | |
| 0.900 | 62.8 | 27.1 | 5.0622 0.7325 | −17.5489 0.0495 | 0.9997 0.9991 | 7.37 × 10 8.86 × 10 | NA NA | |
| 0.800 | 62.9 | 27.0 | 5.0410 0.7373 | −17.7432 0.0345 | 0.9996 0.9992 | 6.41 × 10 5.29 × 10 | NA NA | |
| 0.700 | 51.5 | 30.4 | 4.6601 0.7755 | −20.3361 −0.1303 | 0.9983 0.9998 | 2.40 × 10 1.31 × 10 | NA NA | |
| 0.600 | 67.1 | 25.8 | 5.2604 0.7395 | −18.3228 0.0508 | 0.9998 0.9989 | 6.92 × 10 1.91 × 10 | NA NA | |
| 0.500 | 68.9 | 24.9 | 5.3317 0.7872 | −21.0379 −0.0717 | 0.9994 0.9992 | 9.86 × 10 4.53 × 10 | NA NA | |
| 0.400 | 64.6 | 26.0 | 5.0739 0.7787 | −20.2307 −0.0785 | 0.9991 0.9995 | 3.18 × 10 5.39 × 10 | NA NA | |
| 0.300 | 72.5 | 24.1 | 5.4448 0.7569 | −18.6035 0.0331 | 0.9996 0.9990 | 1.09 × 10 2.74 × 10 | NA NA | |
| 0.201 | 68.0 | 25.1 | 5.3058 0.7854 | −21.1099 −0.0725 | 0.9994 0.9992 | 1.25 × 10 2.56 × 10 | NA NA | |
| 0.101 | 68.5 | 25.0 | 5.3330 0.7857 | −21.2127 −0.0709 | 0.9994 0.9991 | 1.40 × 10 9.34 × 10 | NA NA | |
| - | 129.5 | 12.7 | NA NA | NA NA | NA NA | NA NA | NA NA | |
| - | 131.3 | 12.4 | NA NA | NA NA | NA NA | NA NA | NA NA |
| Matrix | Tablet Desirability | -Value | -Value | Slope | Intercept | p-Value Slope | p-Value Intercept | |
|---|---|---|---|---|---|---|---|---|
| 1.000 | 5.8 | 68.8 | 5.7043 0.7971 | −22.1516 −0.0583 | 0.9999 0.9980 | 7.64 × 10 1.38 × 10 | 2.92 × 10 NA | |
| 1.000 | 5.7 | 69.1 | 5.7087 0.7966 | −22.1438 −0.0567 | 0.9999 0.9980 | 7.20 × 10 1.61 × 10 | 2.97 × 10 NA | |
| 0.928 | 6.5 | 66.3 | 5.6680 0.8111 | −23.0211 −0.1025 | 0.9998 0.9983 | 8.53 × 10 1.30 × 10 | 1.22 × 10 NA | |
| 0.975 | 6.3 | 67.0 | 5.6823 0.8098 | −23.0180 −0.0980 | 0.9998 0.9983 | 9.93 × 10 1.85 × 10 | 1.20 × 10 NA | |
| 0.998 | 6.5 | 66.8 | 5.6632 0.7917 | −21.9957 −0.0508 | 0.9999 0.9978 | 7.98 × 10 1.08 × 10 | 2.90 × 10 NA | |
| 1.000 | 6.3 | 67.3 | 5.6875 0.7991 | −22.1820 −0.0649 | 0.9999 0.9980 | 9.38 × 10 8.23 × 10 | 2.75 × 10 NA | |
| 1.000 | 6.1 | 67.8 | 5.6941 0.7996 | −22.2315 −0.0656 | 0.9999 0.9980 | 8.69 × 10 7.00 × 10 | 2.28 × 10 NA | |
| 1.000 | 6.1 | 67.4 | 5.7011 0.8119 | −23.1813 −0.1020 | 0.9998 0.9982 | 7.91 × 10 1.52 × 10 | 6.73 × 10 NA | |
| 1.000 | 6.6 | 65.9 | 5.6849 0.8141 | −23.1965 −0.1087 | 0.9998 0.9981 | 9.65 × 10 1.55 × 10 | 6.33 × 10 NA | |
| 1.000 | 6.6 | 66.1 | 5.6917 0.8138 | −23.2188 −0.1075 | 0.9998 0.9982 | 8.91 × 10 1.31 × 10 | 6.01 × 10 NA | |
| 0.900 | 9.5 | 58.2 | 6.0098 0.9190 | −30.3709 −0.3713 | 0.9998 0.9971 | 1.49 × 10 1.37 × 10 | NA NA | |
| 0.800 | 7.2 | 64.3 | 5.6312 0.8146 | −23.0534 −0.1147 | 0.9997 0.9984 | 5.01 × 10 2.82 × 10 | 1.34 × 10 NA | |
| 0.700 | 1.8 | 86.8 | 6.0766 0.8091 | −22.5060 −0.0438 | 0.9998 0.9979 | 3.23 × 10 2.31 × 10 | NA NA | |
| 0.600 | 3.1 | 79.8 | 5.7914 0.7993 | −21.3596 −0.0444 | 0.9998 0.9980 | 1.57 × 10 1.39 × 10 | 3.17 × 10 NA | |
| 0.500 | 6.0 | 68.2 | 5.8670 0.8364 | −24.5041 −0.1474 | 0.9999 0.9971 | 1.92 × 10 5.34 × 10 | 1.45 × 10 NA | |
| 0.400 | 6.7 | 65.1 | 6.0149 0.8676 | −27.8541 −0.2317 | 0.9999 0.9972 | 1.42 × 10 1.38 × 10 | NA NA | |
| 0.301 | 5.3 | 70.5 | 6.4250 0.8283 | −24.3255 −0.0636 | 0.9998 0.9979 | 3.57 × 10 4.19 × 10 | NA NA | |
| 0.201 | 6.2 | 67.1 | 6.5041 0.8330 | −24.4633 −0.0649 | 0.9998 0.9981 | 3.22 × 10 6.95 × 10 | NA NA | |
| 0.102 | 3.0 | 81.0 | 5.7810 0.7968 | −21.2583 −0.0392 | 0.9997 0.9983 | 2.07 × 10 8.44 × 10 | 5.01 × 10 NA | |
| - | 46.0 | 20.3 | NA NA | NA NA | NA NA | NA NA | NA NA | |
| - | 45.8 | 20.4 | NA NA | NA NA | NA NA | NA NA | NA NA |
References
- FDA. Guidance for Industry PAT—A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance. Available online: https://www.fda.gov/media/71012/download (accessed on 12 May 2020).
- ICH. Ich Harmonised Tripartite Guideline Pharmaceutical Development Q8(R2). Available online: https://www.ich.org/page/quality-guidelines (accessed on 12 May 2020).
- Yu, L.X.; Amidon, G.; Khan, M.A.; Hoag, S.W.; Polli, J.; Raju, G.K.; Woodcock, J. Understanding pharmaceutical quality by design. AAPS J. 2014, 16, 771–783. [Google Scholar] [CrossRef] [Green Version]
- Leuenberger, H.; Leuenberger, M.N. Impact of the digital revolution on the future of pharmaceutical formulation science. Eur. J. Pharm. Sci. 2016, 87, 100–111. [Google Scholar] [CrossRef]
- Uebbing, L.; Klumpp, L.; Webster, G.K.; Löbenberg, R. Justification of disintegration testing beyond current FDA criteria using in vitro and in silico models. Drug Des. Dev. Ther. 2017, 11, 1163–1174. [Google Scholar] [CrossRef] [Green Version]
- Puchkov, M.; Tschirky, D.; Leuenberger, H. 7-3-D cellular automata in computer-aided design of pharmaceutical formulations: Mathematical concept and F-CAD software. In Formulation Tools for Pharmaceutical Development; Aguilar, J.E., Ed.; Woodhead Publishing: Shaxton, UK, 2013; pp. 155–201. [Google Scholar] [CrossRef]
- Wolfram, S. A New Kind of Science; Wolfram Media Inc.: Champaign, IL, USA, 2002; p. 1197. [Google Scholar]
- Kimura, G.; Puchkov, M.; Leuenberger, H. An attempt to calculate in silico disintegration time of tablets containing mefenamic acid, a low water-soluble drug. J. Pharm. Sci. 2013, 102, 2166–2178. [Google Scholar] [CrossRef]
- Bollmann, S.; Kleinebudde, P. Evaluation of different pre-processing methods of X-ray micro computed tomography images. Powder Technol. 2021, 381, 539–550. [Google Scholar] [CrossRef]
- Bollmann, S.; Kleinebudde, P. Evaluation of different segmentation methods of X-ray micro computed tomography images. Int. J. Pharm. 2021, 120880. [Google Scholar] [CrossRef] [PubMed]
- Bollmann, S.; Kleinebudde, P. Predictive selection rule of favourable image processing methods for X-ray micro-computed tomography images of tablets. IJP 2021. submitted. [Google Scholar]
- Costa, P.; Lobo, J.M.S. Modeling and comparison of dissolution profiles. Eur. J. Pharm. Sci. 2001, 13, 123–133. [Google Scholar] [CrossRef]
- Center for Drug Evaluation and Research (CDER). Guidance for Industry Immediate Release Solid Oral Dosage Forms Scale-Up and Postapproval Changes: Chemistry, Manufacturing, and Controls, In Vitro Dissolution Testing and In Vivo Bioequivalence Documentation; CMC 5; FDA: Silver Spring, MD, USA, 1995.
- CPMP. Note For Guidance on Quality of Modified Release Products: A. Oral Dosage Forms; B. Transdermal Dosage Forms; Section I (Quality); The European Agency for the Evaluation of Medicinal Products: London, UK, 1999.
- Higuchi, T. Rate of Release of Medicaments from Ointment Bases Containing Drugs in Suspension. J. Pharm. Sci. 1961, 50, 874–875. [Google Scholar] [CrossRef]
- Higuchi, T. Mechanism of sustained-action medication. Theoretical analysis of rate of release of solid drugs dispersed in solid matrices. J. Pharm. Sci. 1963, 52, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Korsmeyer, R.W.; Gurny, R.; Doelker, E.; Buri, P.; Peppas, N.A. Mechanisms of solute release from porous hydrophilic polymers. Int. J. Pharm. 1983, 15, 25–35. [Google Scholar] [CrossRef]
- Peppas, N. Analysis of Fickian and non-Fickian drug release from polymers. Pharm. Acta Helv. 1985, 60, 110–111. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Burri, O. Parallel Histogram Matching. Available online: https://gist.github.com/lacan/3de16eb24f954399b763445070fe4bfc (accessed on 3 March 2020).
- Miura, K. Bleach Correction. Available online: https://raw.githubusercontent.com/fiji/CorrectBleach/CorrectBleach_-2.0.2/src/main/java/emblcmci/BleachCorrection.java (accessed on 3 March 2020).
- Miura, K. Histogram Matching. Available online: https://github.com/fiji/CorrectBleach/blob/CorrectBleach_-2.0.2/src/main/java/emblcmci/BleachCorrection_MH.java (accessed on 3 March 2020).
- Münch, B. Remove Background. Available online: https://imagej.net/Xlib (accessed on 5 March 2020).
- Brocher, J. Pseudo Flat Field Correction. Available online: https://github.com/biovoxxel/BioVoxxel_Toolbox-old-/blob/master/Pseudo_flat_field_correction.java (accessed on 5 March 2020).
- Buades, A.; Coll, B.; Morel, J.M. Non-local means denoising. Image Process. Line 2011, 1, 208–212. [Google Scholar] [CrossRef] [Green Version]
- Behnel, P.; Wagner, T. Non-local Means Denoising. Available online: https://raw.githubusercontent.com/thorstenwagner/ij-nl-means/master/src/main/java/de/biomedical_imaging/ij/nlMeansPlugin/NLMeansDenoising_.java (accessed on 5 March 2020).
- Rueden, C. Gamma. Available online: https://github.com/imagej/ImageJA/blob/232620e0a3b0fd33bb22083aaadb2c26b1a31fc6/src/main/java/ij/plugin/filter/ImageMath.java (accessed on 5 March 2020).
- Schindelin, J. Enhance Contrast. Available online: https://github.com/imagej/ImageJA/blob/232620e0a3b0fd33bb22083aaadb2c26b1a31fc6/src/main/java/ij/plugin/ContrastEnhancer.java, (accessed on 5 March 2020).
- Sacha, J. k-Means Clustering. Available online: https://github.com/ij-plugins/ijp-toolkit/blob/master/src/main/java/net/sf/ij_plugins/clustering/KMeansClusteringPlugin.java (accessed on 5 March 2020).
- Jain, A.K. Data clustering: 50 years beyond K-means. Pattern Recognit. Lett. 2010, 31, 651–666. [Google Scholar] [CrossRef]
- Ridler, T.C. Picture Thresholding Using an Iterative Selection Method. IEEE Trans. Syst. Man Cybern. 1978, 8, 630–632. [Google Scholar] [CrossRef]
- Huang, L.K.; Wang, M.J.J. Image thresholding by minimizing the measures of fuzziness. Pattern Recognit. 1995, 28, 41–51. [Google Scholar] [CrossRef]
- Prewitt, J.M.; Mendelsohn, M.L. The analysis of cell images. Ann. N. Y. Acad. Sci. 1966, 128, 1035–1053. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Lee, C.K. Minimum cross entropy thresholding. Pattern Recognit. 1993, 26, 617–625. [Google Scholar] [CrossRef]
- Li, C.H.; Tam, P.K.S. An iterative algorithm for minimum cross entropy thresholding. Pattern Recognit. Lett. 1998, 19, 771–776. [Google Scholar] [CrossRef]
- Sezgin, M.; Sankur, B. Survey over image thresholding techniques and quantitative performance evaluation. J. Electron. Imaging 2004, 13, 146–165. [Google Scholar]
- Kapur, J.N.; Sahoo, P.K.; Wong, A.K.C. A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vis. Graph. Image Process. 1985, 29, 273–285. [Google Scholar] [CrossRef]
- Glasbey, C.A. An Analysis of Histogram-Based Thresholding Algorithms. CVGIP Graph. Model. Image Process. 1993, 55, 532–537. [Google Scholar] [CrossRef]
- Kittler, J.; Illingworth, J. Minimum error thresholding. Pattern Recognit. 1986, 19, 41–47. [Google Scholar] [CrossRef]
- Tsai, W.H. Moment-preserving thresolding: A new approach. Comput. Vis. Graph. Image Process. 1985, 29, 377–393. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. Automatica 1975, 11, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Doyle, W. Operations useful for similarity-invariant pattern recognition. J. ACM (JACM) 1962, 9, 259–267. [Google Scholar] [CrossRef]
- Zack, G.W.; Rogers, W.E.; Latt, S.A. Automatic measurement of sister chromatid exchange frequency. J. Histochem. Cytochem. 1977, 25, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.C.; Chang, F.J.; Chang, S. A new criterion for automatic multilevel thresholding. IEEE Trans. Image Process. 1995, 4, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Rueden, C. Threshold. Available online: https://github.com/fiji/Auto_Threshold/blob/master/src/main/java/fiji/threshold/Auto_Threshold.java (accessed on 11 February 2021).
- Korson, L.; Drost-Hansen, W.; Millero, F.J. Viscosity of water at various temperatures. J. Phys. Chem. 1969, 73, 34–39. [Google Scholar] [CrossRef]
- Hamed, R.; Alnadi, S.H.; Awadallah, A. The Effect of Enzymes and Sodium Lauryl Sulfate on the Surface Tension of Dissolution Media: Toward Understanding the Solubility and Dissolution of Carvedilol. AAPS Pharm. Sci. Tech. 2020, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Anaconda-Inc. Anaconda Software Distribution. Available online: https://docs.anaconda.com/ (accessed on 5 March 2020).
- Moore, J.W.; Flanner, H.H. Mathematical comparison of dissolution profiles. Pharm. Technol. 1996, 20, 64–74. [Google Scholar]
- Sachs, L. Applied Statistics; Springer: Berlin/Heidelberg, Germany, 1984; p. 417. [Google Scholar]
- Hedderich, J.; Sachs, L. Angewandte Statistik; Springer: Berlin/Heidelberg, Germany, 2020; pp. 812–814. [Google Scholar]








| Name | Tablet | Pathway | Tablet Desirability | API | Excipient |
|---|---|---|---|---|---|
| mg | mg | ||||
| real | T25E75-01 | - | - | 89.8 | 320.4 |
| B1 | T25E75-01 | Sta/RB/UC/EC18/G1.7/KM(9) | 1.000 | 88.6 | 316.2 |
| B4 | T25E75-01 | Ref/PFF/UC/EC11/G2.2/KM(11) | 1.000 | 89.5 | 322.6 |
| D | T25E75-01 | - | - | 88.6 | 316.2 |
| G | T25E75-01 | - | - | 88.6 | 316.2 |
| real | T25E75-03 | - | - | 86.9 | 329.5 |
| B1 | T25E75-03 | Sta/RB/UC/EC18/G1.7/KM(9) | 0.982 | 90.0 | 329.0 |
| B2 | T25E75-03 | Sta/RB/UC/G1.3/EC14/KM(7) | 0.991 | 89.3 | 329.6 |
| B3 | T25E75-03 | Sta/UC/UC/EC13/G2.7/KM(12) | 0.958 | 91.1 | 327.6 |
| B4 | T25E75-03 | Ref/PFF/UC/EC11/G2.2/KM(11) | 0.946 | 91.5 | 327.5 |
| C1 | T25E75-03 | Sta/RB/UC/EC17/G1.5/KM(12) | 0.823 | 78.6 | 329.1 |
| C2 | T25E75-03 | Sta/UC/UC/EC13/G2.5/KM(12) | 0.904 | 93.0 | 326.2 |
| API1 | T25E75-03 | Ref/PFF/UC/G3.3/EC13/KM(12) | 0.985 | 84.1 | 332.5 |
| API2 | T25E75-03 | Ref/PFF/UC/EC16/G3.7/KM(12) | 0.789 | 96.3 | 322.4 |
| T1 | T25E75-03 | Ref/RB/NLM/G1.9/EC18/KM(9) | 1.000 | 86.2 | 326.6 |
| T2 | T25E75-03 | Sta/RB/UC/EC18/G1.7/KM(10) | 1.000 | 86.3 | 326.6 |
| T3 | T25E75-03 | Ref/RB/NLM/G1.9/EC18/KM(11) | 0.902 | 80.6 | 325.2 |
| T4 | T25E75-03 | Ref/PFF/NLM/EC15/G2.7/KM(12) | 0.801 | 96.1 | 323.8 |
| T5 | T25E75-03 | Ref/PFF/UC/EC11/G2.3/KM(10) | 0.700 | 98.4 | 322.0 |
| T6 | T25E75-03 | Ref/UC/NLM/EC2/G2.3/MaE-iB//MaE | 0.600 | 98.2 | 305.8 |
| T7 | T25E75-03 | Ref/UC/NLM/ECequalize/G2.1/KM(11) | 0.506 | 73.3 | 329.2 |
| T8 | T25E75-03 | Ref/UC/NLM/EC2/G2.3/MaE//MiE | 0.405 | 98.2 | 286.5 |
| T9 | T25E75-03 | Ref/UC/NLM/EC2/G2.3/MaE//Y-iB | 0.326 | 98.2 | 280.8 |
| T10 | T25E75-03 | Ref/UC/NLM/ECequalize/G2.3/KM(6) | 0.202 | 100.9 | 280.9 |
| T11 | T25E75-03 | Ref/PFF/NLM/ECequalize/G2.1/KM(4) | 0.127 | 73.0 | 275.5 |
| D | T25E75-03 | - | - | 88.6 | 316.2 |
| G | T25E75-03 | - | - | 88.6 | 316.2 |
| real | T25E75-05 | - | - | 86.1 | 331.4 |
| B1 | T25E75-05 | Sta/RB/UC/EC18/G1.7/KM(9) | 0.955 | 90.4 | 330.3 |
| B4 | T25E75-05 | Ref/PFF/UC/EC11/G2.2/KM(11) | 0.928 | 91.4 | 329.4 |
| D | T25E75-05 | - | - | 88.6 | 316.2 |
| G | T25E75-05 | - | - | 88.6 | 316.2 |
| Parameter | Value |
|---|---|
| general | |
| cube-X, cube-Y, cube-Z | image size |
| unitcube | 0.028 mm |
| iterations | 43,200 |
| timeformat | seconds |
| sampling interval | 600 |
| mixing parameter | 0.05 |
| theophylline | |
| identification number | 1 |
| density | 1447.2 kg/m³ |
| solubility | 10.09 mg/mL |
| molar mass | 180.16 Da |
| ethyl cellulose | |
| identification number | 31 |
| density | 1139.9 kg/m³ |
| solubility | 0.00 mg/mL |
| molar mass | 75,000.0 Da |
| contact angle | 89° |
| porosity | calculated by Equation (2) (%) |
| pore size | 0.5 m |
| phosphate buffer | |
| identification number | 200 |
| density | 1007.0 kg/m³ |
| viscosity | 0.0006865 Pa s |
| tension | 67.9 mN/m |
| air | |
| identification number | 0 |
| Matrix | Tablet Desirability | -Value | -Value | Slope | Intercept | p-Value Slope | p-Value Intercept | |
|---|---|---|---|---|---|---|---|---|
| real data | - | - | - | 1.7869 0.4802 | 1.0926 0.3172 | 0.9996 0.9990 | - | - |
| 0.985 | 11.7 | 66.1 | 2.5605 0.6509 | −10.7717 −0.0910 | 0.9993 0.9983 | 2.94 × 10 2.14 × 10 | NA NA | |
| 0.789 | 20.3 | 54.2 | 2.7863 0.6512 | −11.0816 −0.0478 | 0.9992 0.9985 | 4.70 × 10 1.06 × 10 | NA NA | |
| 0.823 | 22.6 | 55.4 | 2.1661 0.7450 | −15.2112 −0.4924 | 0.9959 0.9982 | 3.55 × 10 2.51 × 10 | NA NA | |
| 0.904 | 9.8 | 70.9 | 2.6728 0.7356 | −16.739 −0.3548 | 0.9989 0.9997 | 9.86 × 10 2.05 × 10 | NA NA | |
| 0.982 | 10.3 | 69.4 | 2.5019 0.7382 | −16.1765 −0.3962 | 0.9986 0.9997 | 6.51 × 10 1.37 × 10 | NA NA | |
| 0.991 | 10.6 | 68.9 | 2.4955 0.7384 | −16.3043 −0.3996 | 0.9987 0.9997 | 9.48 × 10 2.30 × 10 | NA NA | |
| 0.958 | 9.5 | 71.5 | 2.6392 0.7369 | −16.7893 −0.3667 | 0.9989 0.9997 | 4.32 × 10 2.74 × 10 | NA NA | |
| 0.946 | 17.0 | 57.9 | 2.7147 0.6526 | −11.1982 −0.0675 | 0.9994 0.9984 | 6.12 × 10 2.32 × 10 | NA NA | |
| 1.000 | 12.5 | 63.9 | 2.7689 0.7000 | −15.0743 −0.2217 | 0.9984 0.9995 | 1.85 × 10 3.01 × 10 | NA NA | |
| 1.000 | 14.0 | 64.3 | 2.3850 0.7458 | −16.0278 −0.4445 | 0.9988 0.9992 | 9.05 × 10 5.35 × 10 | NA NA | |
| 0.902 | 8.7 | 72.2 | 2.5916 0.6875 | −13.9408 −0.215 | 0.9967 0.9993 | 1.17 × 10 4.16 × 10 | NA NA | |
| 0.801 | 40.4 | 40.9 | 3.0837 0.6132 | −8.3566 0.1323 | 0.9990 0.9989 | 2.12 × 10 3.12 × 10 | NA NA | |
| 0.700 | 21.8 | 52.8 | 2.8241 0.6520 | −11.1975 −0.0436 | 0.9993 0.9985 | 3.96 × 10 1.05 × 10 | NA NA | |
| 0.600 | 30.7 | 45.6 | 3.0188 0.6516 | −11.5268 −0.0083 | 0.9985 0.9995 | 1.03 × 10 1.03 × 10 | NA NA | |
| 0.506 | 7.7 | 75.5 | 2.5034 0.6785 | −13.2446 −0.2039 | 0.9948 0.9981 | 1.44 × 10 2.86 × 10 | NA NA | |
| 0.405 | 48.7 | 36.8 | 3.3312 0.6346 | −10.0969 0.1038 | 0.9996 0.9997 | 2.39 × 10 8.02 × 10 | NA NA | |
| 0.326 | 58.0 | 33.2 | 3.5779 0.6480 | −11.2144 0.1001 | 0.9997 0.9997 | 2.04 × 10 3.70 × 10 | NA NA | |
| 0.202 | 57.7 | 33.3 | 3.5908 0.6508 | −11.4484 0.0928 | 0.9996 0.9997 | 2.42 × 10 1.00 × 10 | NA NA | |
| 0.127 | 81.2 | 26.5 | 4.0747 0.6462 | −11.1658 0.1777 | 0.9992 0.9998 | 3.76 × 10 7.30 × 10 | NA NA | |
| - | 30.0 | 44.7 | NA NA | NA NA | 0.1855 0.3603 | 4.12 × 10 1.72 × 10 | NA NA | |
| - | 125.4 | 18.4 | 3.1517 0.2779 | 17.4812 1.1372 | 0.9975 0.9913 | 1.08 × 10 4.85 × 10 | NA NA | |
| - | 111.3 | 21.2 | 5.9146 0.3874 | 11.9812 1.0768 | 0.9559 0.9687 | 2.15 × 10 5.38 × 10 | NA NA | |
| - | 146.5 | 15.3 | 5.2757 0.3997 | 9.7346 0.9971 | 0.9793 0.9841 | 1.16 × 10 5.95 × 10 | NA NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bollmann, S.; Kleinebudde, P. A New Validation Methodology for In Silico Tools Based on X-ray Computed Tomography Images of Tablets and a Performance Analysis of One Tool. Pharmaceutics 2021, 13, 1488. https://doi.org/10.3390/pharmaceutics13091488
Bollmann S, Kleinebudde P. A New Validation Methodology for In Silico Tools Based on X-ray Computed Tomography Images of Tablets and a Performance Analysis of One Tool. Pharmaceutics. 2021; 13(9):1488. https://doi.org/10.3390/pharmaceutics13091488
Chicago/Turabian StyleBollmann, Sebastian, and Peter Kleinebudde. 2021. "A New Validation Methodology for In Silico Tools Based on X-ray Computed Tomography Images of Tablets and a Performance Analysis of One Tool" Pharmaceutics 13, no. 9: 1488. https://doi.org/10.3390/pharmaceutics13091488
APA StyleBollmann, S., & Kleinebudde, P. (2021). A New Validation Methodology for In Silico Tools Based on X-ray Computed Tomography Images of Tablets and a Performance Analysis of One Tool. Pharmaceutics, 13(9), 1488. https://doi.org/10.3390/pharmaceutics13091488