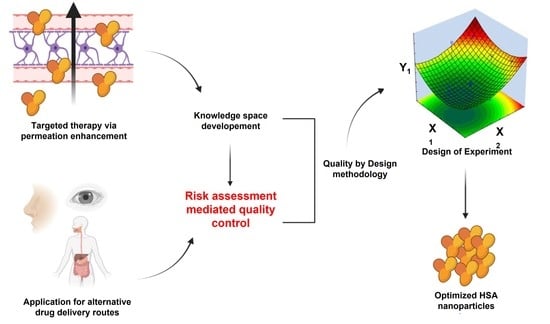
Risk-Assessment-Based Optimization Favours the Development of Albumin Nanoparticles with Proper Characteristics Prior to Drug Loading
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Definition of the Quality Target Product Profile (QTPP)
2.3. Determination of CQA, CMA, and CPP Elements
2.4. Initial Risk Assessment
2.5. Screening Study Using Plackett–Burman Design
2.6. Preparation of HSA Nanoparticles
2.7. Optimizing the Rapid Coacervation Method for HSA Nanoparticle Formation
2.8. Average Hydrodynamic Diameter, Polydispersity Index and Zeta Potential Determination
2.9. Determination a Nanoparticle Yield after Coacervation
2.10. Raman Spectroscopy
3. Results
3.1. Determination of Quality Target Product Profiles
3.2. Initial Risk Assessment on the Rapid Coacervation Method for HSA Nanoparticle Development
3.3. Screening of Optimizable Factors via Plackett–Burman Design
3.3.1. Screening the Effect of Process Variables on Average Hydrodynamic Diameter
3.3.2. Screening the Effect of Process Variables on the Polydispersity Index
3.3.3. Screening the Effect of Process Variables on Zeta Potential
3.4. Optimization of HSA Nanoparticles with Box–Behnken Experimental Design
3.4.1. Influence of Process Variables on the Z-Average (Box–Behnken Design)
3.4.2. Influence of Process Variables on the PdI (Box–Behnken Design)
3.4.3. Influence of Process Variables on the Zeta Potential (Box–Behnken Design)
3.5. Confirmation Test of the Model
3.6. Raman Spectroscopic Structural Investigation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schweizer, L.; Dingermann, T. Introduction: Trends and Developments in the Pharmaceutical and Life Sciences Industry. In Advances in Pharma Business Management and Research: Volume 1; Schweizer, L., Dingermann, T., Russe, O.Q., Jansen, C., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–5. ISBN 978-3-030-35918-8. [Google Scholar]
- Sipos, B.; Katona, G.; Csóka, I. A systematic, knowledge space-based proposal on quality by design-driven polymeric micelle development. Pharmaceutics 2021, 13, 702. [Google Scholar] [CrossRef] [PubMed]
- Khanna, I. Drug discovery in pharmaceutical industry: Productivity challenges and trends. Drug Discov. Today 2012, 17, 1088–1102. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, H.; Yang, T.; Li, T.; Zhang, M.; Ke, H.; Ding, D.; Deng, Y.; Chen, H. Serum protein-based nanoparticles for cancer diagnosis and treatment. J. Control. Release 2021, 329, 997–1022. [Google Scholar] [CrossRef] [PubMed]
- Lomis, N.; Westfall, S.; Farahdel, L.; Malhotra, M.; Shum-Tim, D.; Prakash, S. Human serum albumin nanoparticles for use in cancer drug delivery: Process optimization and in vitro characterization. Nanomaterials 2016, 6, 116. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, A.; Azoia, N.G.; Gomes, A.C.; Cavaco-Paulo, A. Albumin-Based Nanodevices as Drug Carriers. Curr. Pharm. Des. 2016, 22, 1371–1390. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jiao, Y.; Wang, Y.; Zhou, C.; Zhang, Z. Polysaccharides-based nanoparticles as drug delivery systems. Adv. Drug Deliv. Rev. 2008, 60, 1650–1662. [Google Scholar] [CrossRef] [PubMed]
- Elzoghby, A.O.; Samy, W.M.; Elgindy, N.A. Albumin-based nanoparticles as potential controlled release drug delivery systems. J. Control. Release 2012, 157, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Hashem, L.; Swedrowska, M.; Vllasaliu, D. Intestinal uptake and transport of albumin nanoparticles: Potential for oral delivery. Nanomedicine 2018, 13, 1255–1265. [Google Scholar] [CrossRef]
- An, F.F.; Zhang, X.H. Strategies for preparing albumin-based nanoparticles for multifunctional bioimaging and drug delivery. Theranostics 2017, 7, 3667–3689. [Google Scholar] [CrossRef]
- Hersh, D.S.; Wadajkar, A.S.; Roberts, N.; Perez, J.G.; Connolly, N.P.; Frenkel, V.; Winkles, J.A.; Woodworth, G.F.; Kim, A.J. Evolving Drug Delivery Strategies to Overcome the Blood Brain Barrier. Curr. Pharm. Des. 2016, 22, 1177–1193. [Google Scholar] [CrossRef] [Green Version]
- Wong, L.R.; Ho, P.C. Role of serum albumin as a nanoparticulate carrier for nose-to-brain delivery of R-flurbiprofen: Implications for the treatment of Alzheimer’s disease. J. Pharm. Pharmacol. 2018, 70, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Tiozzo Fasiolo, L.; Manniello, M.D.; Bortolotti, F.; Buttini, F.; Rossi, A.; Sonvico, F.; Colombo, P.; Valsami, G.; Colombo, G.; Russo, P. Anti-inflammatory flurbiprofen nasal powders for nose-to-brain delivery in Alzheimer’s disease. J. Drug Target. 2019, 27, 984–994. [Google Scholar] [CrossRef] [PubMed]
- Katona, G.; Balogh, G.T.; Dargó, G.; Gáspár, R.; Márki, Á.; Ducza, E.; Sztojkov-Ivanov, A.; Tömösi, F.; Kecskeméti, G.; Janáky, T.; et al. Development of meloxicam-human serum albumin nanoparticles for nose-to-brain delivery via application of a quality by design approach. Pharmaceutics 2020, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Morishita, M.; Peppas, N.A. Is the oral route possible for peptide and protein drug delivery? Drug Discov. Today 2006, 11, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Niknejad, H.; Mahmoudzadeh, R. Comparison of different crosslinking methods for preparation of docetaxel-loaded albumin nanoparticles. Iran. J. Pharm. Res. 2015, 14, 385–394. [Google Scholar]
- Oh, H.N.; Yoo, D.; Park, S.; Lee, S.; Kim, W.K. Developmental neurotoxicity induced by glutaraldehyde in neuron/astrocyte co-cultured cells and zebrafish. Ecotoxicol. Environ. Saf. 2022, 242, 113891. [Google Scholar] [CrossRef]
- Jarquín-Yáñez, K.; Arenas-Alatorre, J.; Piñón-Zárate, G.; Arellano-Olivares, R.M.; Herrera-Enríquez, M.; Hernández-Téllez, B.; Castell-Rodríguez, A.E. Structural effect of different EDC crosslinker concentration in gelatin-hyaluronic acid scaffolds. J. Bioeng. Biomed. Sci. 2016, 6, 182. [Google Scholar] [CrossRef]
- Sunoqrot, S.; Alsadi, A.; Tarawneh, O.; Hamed, R. Polymer type and molecular weight dictate the encapsulation efficiency and release of Quercetin from polymeric micelles. Colloid. Polym. Sci. 2017, 295, 2051–2059. [Google Scholar] [CrossRef]
- Pallagi, E.; Jójárt-Laczkovich, O.; Németh, Z.; Szabó-Révész, P.; Csóka, I. Application of the QbD-based approach in the early development of liposomes for nasal administration. Int. J.Pharm. 2019, 562, 11–22. [Google Scholar] [CrossRef]
- Csóka, I.; Pallagi, E.; Paál, T.L. Extension of quality-by-design concept to the early development phase of pharmaceutical R&D processes. Drug Discov. Today 2018, 23, 1340–1343. [Google Scholar] [CrossRef]
- ICH. Quality Risk Management Q9; ICH Harmonised Tripartite Guideline; ICH: Geneva, Switzerland, 2005. [Google Scholar]
- ICH. Pharmaceutical Quality Systems Q10; ICH Harmonised Tripartite Guideline; ICH: Geneva, Switzerland, 2008. [Google Scholar]
- ICH. Pharmaceutical Development Q8; ICH Harmonised Tripartite Guideline; ICH: Geneva, Switzerland, 2009. [Google Scholar]
- Rakotoarisoa, M.; Angelov, B.; Drechsler, M.; Nicolas, V.; Bizien, T.; Gorshkova, Y.E.; Deng, Y.; Angelova, A. Liquid crystalline lipid nanoparticles for combined delivery of curcumin, fish oil and BDNF: In vitro neuro-protective potential in a cellular model of tunicamycin-induced endoplasmic reticulum stress. Smart Mater. Med. 2022, 3, 274–288. [Google Scholar] [CrossRef]
- Analytical Methods Committee. AMCTB No 55. Experimental design and optimisation (4): Plackett–Burman designs. Anal. Methods 2013, 5, 1901–1903. [Google Scholar] [CrossRef] [PubMed]
- Jahanban-Esfahlan, A.; Dastmalchi, S.; Davaran, S. A simple improved desolvation method for the rapid preparation of albumin nanoparticles. Int. J. Biol. Macromol. 2016, 91, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Von Storp, B.; Engel, A.; Boeker, A.; Ploeger, M.; Langer, K. Albumin nanoparticles with predictable size by desolvation procedure. J. Microencapsul. 2012, 29, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Mohammad-Beigi, H.; Shojaosadati, S.A.; Morshedi, D.; Mirzazadeh, N.; Arpanaei, A. The effects of organic solvents on the physicochemical properties of human serum albumin nanoparticles. Iran. J. Biotechnol. 2016, 14, 45–50. [Google Scholar] [CrossRef]
- ICH. Guideline for residual solvents Q3C(R5); ICH Harmonised Tripartite Guideline; ICH: Geneva, Switzerland, 2011. [Google Scholar]
- Sadeghi, R.; Moosavi-Movahedi, A.A.; Emam-Jomeh, Z.; Kalbasi, A.; Razavi, S.H.; Karimi, M.; Kokini, J. The effect of different desolvating agents on BSA nanoparticle properties and encapsulation of curcumin. J. Nanoparticle Res. 2014, 16, 1–14. [Google Scholar] [CrossRef]
- Bae, S.; Ma, K.; Kim, T.H.; Lee, E.S.; Oh, K.T.; Park, E.S.; Lee, K.C.; Youn, Y.S. Doxorubicin-loaded human serum albumin nanoparticles surface-modifiedwith TNF-related apoptosis-inducing ligand and transferrin for targetingmultiple tumor types. Biomaterials 2012, 33, 1536–1546. [Google Scholar] [CrossRef]
- Langer, K.; Balthasar, S.; Vogel, V.; Dinauer, N.; Von Briesen, H.; Schubert, D. Optimization of the preparation process for human serum albumin (HSA) nanoparticles. Int. J. Pharm. 2003, 257, 169–180. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Yang, X.; Zhang, S.; Cui, C. Preparation optimization of bovine serum albumin nanoparticles and its application for siRNA delivery. Drug Des. Devel. Ther. 2021, 15, 1531–1547. [Google Scholar] [CrossRef]
- Hornok, V. Serum albumin nanoparticles: Problems and prospects. Polymers 2021, 13, 3759. [Google Scholar] [CrossRef]
- Sebak, S.; Mirzaei, M.; Malhotra, M.; Kulamarva, A.; Prakash, S. Human serum albumin nanoparticles as an efficient noscapine drug delivery system for potential use in breast cancer: Preparation and in vitro analysis. Int. J. Nanomed. 2010, 5, 525–532. [Google Scholar] [CrossRef] [Green Version]
- Taha, M.S.; Padmakumar, S.; Singh, A.; Amiji, M.M. Critical quality attributes in the development of therapeutic nanomedicines toward clinical translation. Drug Deliv. Transl. Res. 2020, 10, 766–790. [Google Scholar] [CrossRef] [PubMed]
- Jithan, A.V.; Madhavi, K.; Madhavi, M.; Prabhakar, K. Preparation and characterization of albumin nanoparticles encapsulating curcumin intended for the treatment of breast cancer. Int. J. Pharm. Investig. 2011, 1, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Rohiwal, S.S.; Tiwari, A.P.; Verma, G.; Pawar, S.H. Preparation and evaluation of bovine serum albumin nanoparticles for ex vivo colloidal stability in biological media. Colloids. Surf. A: Physicochem. Eng. Asp. 2015, 480, 28–37. [Google Scholar] [CrossRef]
- Füredi, P.; Kovács, K.; Ludányi, K.; Antal, I.; Klebovich, I. Development and characterization of voriconazole loaded nanoparticles for parenteral delivery. Int. J. Pharm. 2016, 510, 159–163. [Google Scholar] [CrossRef]
- David, C.; Foley, S.; Mavon, C.; Enescu, M. Reductive unfolding of serum albumins uncovered by Raman spectroscopy. Biopolym. Orig. Res. Biomol. 2008, 89, 623–634. [Google Scholar] [CrossRef]
- Emin, A.; Hushur, A.; Mamtimin, T. Raman study of mixed solutions of methanol and ethanol. AIP Adv. 2020, 10, 065330. [Google Scholar] [CrossRef]







| Formulation Variables | Code | Levels | |
|---|---|---|---|
| −1 | +1 | ||
| Amount of ethanol (mL) | X1 | 2 | 4 |
| Amount of crosslinker (EDC) (mg) | X2 | 2.5 | 5 |
| Incubation time (h) | X3 | 1 | 3 |
| Concentration of HSA (mg/mL) | X4 | 50 | 75 |
| Ethanol flow rate (mL/min) | X5 | 1 | 2 |
| Stirring speed (rpm) | X6 | 750 | 1500 |
| Concentration of NaCl (w/v%) | X7 | 0 | 0.9 |
| Independent Variables | Code | Levels | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| Amount of ethanol (mL) | X1 | 1 | 2 | 3 |
| Incubation time (h) | X3 | 3 | 4.5 | 6 |
| Concentration of HSA (mg/mL) | X4 | 50 | 62.5 | 75 |
| QTPP Element | Aim | Justification |
|---|---|---|
| Particle size (expressed as average hydrodynamic diameter) | <200 nm | Nanoparticles below 200 nm show increased surface area, which contributes to a highly diffusive area of drug release and permeation tendencies. |
| Particle size distribution (expressed as polydispersity index) | <0.300 | PdI below 0.3 results in a uniform drug release and permeability profile, allowing controllable targeting and drug administration. Uniform particle size prior to drug loading also increases the potential of the formation of uniform drug-bound albumin particles. |
| Zeta potential | >│15 mV│ | Generally speaking, nanoparticles with a zeta potential value above an absolute value of 15 mV (in either charge direction) are considered stable, as the repulsion forces are increased amongst particles. |
| Drug-binding capacity | Capable of binding the required amount of active substance | It depends on the target active substance concentration, the desired administration route, and the chosen dosage form. Generally, it should be as high as possible based on the reachable equilibrium amongst the active substance and albumin. |
| Drug release profile | Depends on the active substance, additional excipients, the administration route, and the expected pharmacokinetic profile. | |
| Drug permeability profile | Depends on the active substance, additional excipients, the administration route, and the expected pharmacokinetic profile. | |
| Process Variables | Response | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Batch ID | X1 (mL) | X2 (mg) | X3 (h) | X4 (mg/mL) | X5 (mL/min) | X6 (rpm) | X7 (w/v%) | Y1 (nm) | Y2 | Y3 (mV) |
| PB1 | 2 | 2.5 | 1 | 75 | 2 | 1500 | 0 | 90 ± 11 | 0.445 ± 0.065 | −18.37 ± 4.65 |
| PB2 | 4 | 2.5 | 1 | 50 | 1 | 1500 | 0.9 | 1828 ± 103 | 0.836 ± 0.044 | −3.37 ± 2.94 |
| PB3 | 2 | 5 | 1 | 50 | 2 | 750 | 0.9 | 194 ± 16 | 0.405 ± 0.169 | −6.04 ± 0.88 |
| PB4 | 4 | 5 | 1 | 75 | 1 | 750 | 0 | 109 ± 19 | 0.401 ± 0.106 | −27.07 ± 3.96 |
| PB5 | 2 | 2.5 | 3 | 75 | 1 | 750 | 0.9 | 181 ± 19 | 0.818 ± 0.052 | −5.25 ± 2.06 |
| PB6 | 4 | 2.5 | 3 | 50 | 2 | 750 | 0 | 116 ± 4 | 0.230 ± 0.033 | −11.00 ± 0.26 |
| PB7 | 2 | 5 | 3 | 50 | 1 | 1500 | 0 | 131 ± 25 | 0.430 ± 0.023 | −10.27 ± 1.31 |
| PB8 | 4 | 5 | 3 | 75 | 2 | 1500 | 0.9 | 233 ± 9 | 0.190 ± 0.086 | −22.03 ± 3.45 |
| Variable | Code | Significance | Effect of Variable on Z-Average |
|---|---|---|---|
| Amount of ethanol (mL) | X1 | ** p < 0.01 | + |
| Amount of crosslinker (EDC) (mg) | X2 | * p < 0.05 | − |
| Incubation time (h) | X3 | * p < 0.05 | − |
| Concentration of HSA (mg/mL) | X4 | ** p < 0.01 | − |
| Ethanol flow rate (mL/min) | X5 | * p < 0.05 | − |
| Stirring speed (rpm) | X6 | ** p < 0.01 | + |
| Concentration of NaCl (w/v%) | X7 | ** p < 0.01 | + |
| Variable | Code | Significance | Effect of Variable on PdI |
|---|---|---|---|
| Amount of ethanol (mL) | X1 | n.s. | − |
| Amount of crosslinker (EDC) (mg) | X2 | * p < 0.05 | − |
| Incubation time (h) | X3 | n.s. | − |
| Concentration of HSA (mg/mL) | X4 | n.s. | + |
| Ethanol flow rate (mL/min) | X5 | ** p < 0.01 | − |
| Stirring speed (rpm) | X6 | n.s. | + |
| Concentration of NaCl (w/v%) | X7 | ** p < 0.01 | + |
| Variable | Code | Significance | Effect of Variable on Colloidal Stability |
|---|---|---|---|
| Amount of ethanol (mL) | X1 | ** p < 0.01 | + |
| Amount of crosslinker (EDC) (mg) | X2 | ** p < 0.01 | + |
| Incubation time (h) | X3 | n.s. | − |
| Concentration of HSA (mg/mL) | X4 | ** p < 0.01 | + |
| Ethanol flow rate (mL/min) | X5 | * p < 0.05 | + |
| Stirring speed (rpm) | X6 | n.s. | + |
| Concentration of NaCl (w/v%) | X7 | ** p < 0.01 | − |
| Process Variables | Response | ||||||
|---|---|---|---|---|---|---|---|
| Batch ID | X1 (mL) | X3 (h) | X4 (mg/mL) | Y1 (nm) | Y2 | Y3 (mV) | Y4 (%) |
| BB1 | 1 | 4.5 | 50 | 485 ± 26 | 0.901 ± 0.019 | −28.9 ± 3.6 | 83.3 ± 3.4 |
| BB2 | 3 | 4.5 | 50 | 117 ± 11 | 0.436 ± 0.181 | −24.5 ± 4.9 | 94.8 ± 2.8 |
| BB3 | 1 | 4.5 | 75 | 489 ± 22 | 0.876 ± 0.107 | −30.5 ± 4.7 | 82.7 ± 4.1 |
| BB4 | 3 | 4.5 | 75 | 144 ± 7 | 0.652 ± 0.112 | −37 ± 3.1 | 98 ± 1.2 |
| BB5 | 1 | 3 | 62.5 | 434 ± 6 | 0.871 ± 0.109 | −26.7 ± 6.3 | 78.2 ± 3.9 |
| BB6 | 3 | 3 | 62.5 | 168 ± 21 | 0.412 ± 0.156 | −35.2 ± 4.5 | 95.6 ± 1.5 |
| BB7 | 1 | 6 | 62.5 | 308 ± 26 | 0.731 ± 0.124 | −30 ± 4.2 | 83.5 ± 2.8 |
| BB8 | 3 | 6 | 62.5 | 111 ± 8 | 0.507 ± 0.161 | −36.4 ± 4.4 | 91.3 ± 3.1 |
| BB9 | 2 | 3 | 50 | 197 ± 21 | 0.453 ± 0.113 | −29.4 ± 2.6 | 88 ± 2.9 |
| BB10 | 2 | 3 | 75 | 186 ± 19 | 0.442 ± 0.062 | −29.5 ± 6.5 | 94.4 ± 2 |
| BB11 | 2 | 6 | 50 | 196 ± 20 | 0.438 ± 0.172 | −30.1 ± 7.4 | 92.6 ± 3.7 |
| BB12 | 2 | 6 | 75 | 117 ± 18 | 0.429 ± 0.185 | −26.7 ± 2.7 | 92.9 ± 2.5 |
| BB13 | 2 | 4.5 | 62.5 | 110 ± 5 | 0.251 ± 0.051 | −29.6 ± 4.6 | 92 ± 1.7 |
| BB14 | 2 | 4.5 | 62.5 | 110 ± 6 | 0.253 ± 0.059 | −30.2 ± 4.1 | 93.9 ± 2.1 |
| BB15 | 2 | 4.5 | 62.5 | 119 ± 4 | 0.247 ± 0.063 | −30.4 ± 4.2 | 92.5 ± 2.2 |
| Variable | Code | Significance | Effect of Variable on Z-Average |
|---|---|---|---|
| Amount of ethanol (mL) | X1 | ** p < 0.01 | − |
| Amount of ethanol (mL) | X12 | * p < 0.05 | − |
| Incubation time (h) | X3 | n.s. | − |
| Incubation time (h) | X32 | n.s. | − |
| Concentration of HSA (mg/mL) | X4 | n.s. | − |
| Concentration of HSA (mg/mL) | X42 | * p < 0.05 | − |
| Variable | Code | Significance | Effect of Variable on PdI |
|---|---|---|---|
| Amount of ethanol (mL) | X1 | ** p < 0.01 | − |
| Amount of ethanol (mL) | X12 | ** p < 0.01 | − |
| Incubation time (h) | X3 | n.s. | − |
| Incubation time (h) | X32 | n.s. | + |
| Concentration of HSA (mg/mL) | X4 | n.s. | + |
| Concentration of HSA (mg/mL) | X42 | ** p < 0.01 | − |
| Variable | Code | Significance | Effect of Variable on Colloidal Stability |
|---|---|---|---|
| Amount of ethanol (mL) | X1 | n.s. | − |
| Amount of ethanol (mL) | X12 | n.s. | − |
| Incubation time (h) | X3 | n.s. | + |
| Incubation time (h) | X32 | n.s. | − |
| Concentration of HSA (mg/mL) | X4 | n.s. | + |
| Concentration of HSA (mg/mL) | X42 | n.s. | + |
| Process Variables | Response | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| X1 (mL) | X2 (mg) | X3 (h) | X4 (mg/mL) | X5 (mL/min) | X6 (rpm) | X7 (w/v%) | Y1 (nm) | Y2 | Y3 (mV) | Y4 (%) | |
| Predicted | 2.5 | 2.5 | 3 | 62.5 | 2 | 750 | 0 | 122 | 0.231 | −28.4 | - |
| Experimental | 119 ± 5 | 0.236 ± 0.051 | −27.2 ± 2.6 | 96.3 ± 1.7 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katona, G.; Sipos, B.; Csóka, I. Risk-Assessment-Based Optimization Favours the Development of Albumin Nanoparticles with Proper Characteristics Prior to Drug Loading. Pharmaceutics 2022, 14, 2036. https://doi.org/10.3390/pharmaceutics14102036
Katona G, Sipos B, Csóka I. Risk-Assessment-Based Optimization Favours the Development of Albumin Nanoparticles with Proper Characteristics Prior to Drug Loading. Pharmaceutics. 2022; 14(10):2036. https://doi.org/10.3390/pharmaceutics14102036
Chicago/Turabian StyleKatona, Gábor, Bence Sipos, and Ildikó Csóka. 2022. "Risk-Assessment-Based Optimization Favours the Development of Albumin Nanoparticles with Proper Characteristics Prior to Drug Loading" Pharmaceutics 14, no. 10: 2036. https://doi.org/10.3390/pharmaceutics14102036
APA StyleKatona, G., Sipos, B., & Csóka, I. (2022). Risk-Assessment-Based Optimization Favours the Development of Albumin Nanoparticles with Proper Characteristics Prior to Drug Loading. Pharmaceutics, 14(10), 2036. https://doi.org/10.3390/pharmaceutics14102036