Genotyping of UGT1A1*80 as an Alternative to UGT1A1*28 Genotyping in Spain
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Linkage Disequilibrium Analyses
3.2. Real Predictive Values for rs887829 Genotyping
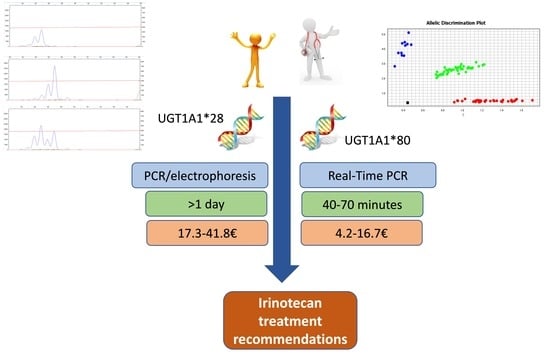
3.3. Comparison of Genotyping Cost and Time to Result
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rowinsky, E.K.; Grochow, L.B.; Ettinger, D.S.; Sartorius, S.E.; Lubejko, B.G.; Chen, T.L.; Rock, M.K.; Donehower, R.C. Phase I and pharmacological study of the novel topoisomerase I inhibitor 7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxycamptothecin (CPT-11) administered as a ninety-minute infusion every 3 weeks. Cancer Res. 1994, 54, 427–436. [Google Scholar] [PubMed]
- Atasilp, C.; Biswas, M.; Jinda, P.; Nuntharadthanaphong, N.; Rachanakul, J.; Hongkaew, Y.; Vanwong, N.; Saokaew, S.; Sukasem, C. Association of UGT1A1*6,*28 or ABCC2 c.3972C>T genetic polymorphisms with irinotecan induced toxicity in Asian cancer patients: Meta-analysis. Clin. Transl. Sci. 2022, 15, 1613–1633. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.S.; Seligson, N.D.; Bottiglieri, S.; Carballido, E.; Del Cueto, A.; Imanirad, I.; Levine, R.; Parker, A.S.; Swain, S.M.; Tillman, E.M.; et al. UGT1A1 Guided Cancer Therapy: Review of the Evidence and Considerations for Clinical Implementation. Cancers 2021, 13, 1566. [Google Scholar] [CrossRef] [PubMed]
- Kamal, S.; Abdelhakam, S.; Ghoraba, D.; Massoud, Y.; Aziz, K.A.; Hassan, H.; Hafez, T.; Abdel Sallam, A. The frequency, clinical course, and health related quality of life in adults with Gilbert’s syndrome: A longitudinal study. BMC Gastroenterol. 2019, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cao, W.; Ding, H.; Liu, T.; Zhou, X.; Wang, M.; Zhong, M.; Zhao, Z.; Xu, Q.; Wang, L. Analysis of UGT1A1*28 genotype and SN-38 pharmacokinetics for irinotecan-based chemotherapy in patients with advanced colorectal cancer: Results from a multicenter, retrospective study in Shanghai. J. Cancer Res. Clin. Oncol. 2013, 139, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Gammal, R.S.; Court, M.H.; Haidar, C.E.; Iwuchukwu, O.F.; Gaur, A.H.; Alvarellos, M.; Guillemette, C.; Lennox, J.L.; Whirl-Carrillo, M.; Brummel, S.S.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing. Clin. Pharmacol. Ther. 2016, 99, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; Rongen, G.A.P.J.M.; van Schaik, R.H.N.; Schalekamp, T.; Touw, D.J.; et al. Pharmacogenetics: Form bench to byte—An update of guidelines. Clin. Pharmacol. Ther. 2011, 89, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Quaranta, S.; Thomas, F. Pharmacogenetics of anti-cancer drugs: State of the art and implementation—Recommendations of the French National Network of Pharmacogenetics. Therapie 2017, 72, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Cortejoso, L.; López-Fernández, L.A. Pharmacogenetic markers of toxicity for chemotherapy in colorectal cancer patients. Pharmacogenomics 2012, 13, 1173–1191. [Google Scholar] [CrossRef] [PubMed]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef] [PubMed]
- Innocenti, F.; Undevia, S.D.; Iyer, L.; Xian Chen, P.; Das, S.; Kocherginsky, M.; Karrison, T.; Janisch, L.; Ramírez, J.; Rudin, C.M.; et al. Genetic Variants in the UDP-glucuronosyltransferase 1A1 Gene Predict the Risk of Severe Neutropenia of Irinotecan. J. Clin. Oncol. 2004, 22, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Daprà, V.; Alliaudi, C.; Galliano, I.; Dini, M.; Lo Curcio, G.; Calvi, C.; Archetti, M.; Gavatorta, M.; Bergallo, M. TaqMan real time PCR for the Detection of the Gilbert’s Syndrome Markers UGT1A1*28; UGT1A1*36 and UGT1A1*37. Mol. Biol. Rep. 2021, 48, 4953–4959. [Google Scholar] [CrossRef] [PubMed]
- Ehmer, U.; Lankisch, T.O.; Erichsen, T.J.; Kalthoff, S.; Freiberg, N.; Wehmeier, M.; Manns, M.P.; Strassburg, C.P. Rapid allelic discrimination by TaqMan PCR for the detection of the Gilbert’s syndrome marker UGT1A1*28. J. Mol. Diagn. 2008, 10, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Hulshof, E.C.; Deenen, M.J.; Guchelaar, H.-J.; Gelderblom, H. Pre-therapeutic UGT1A1 genotyping to reduce the risk of irinotecan-induced severe toxicity: Ready for prime time. Eur. J. Cancer 2020, 141, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Karas, S.; Innocenti, F. All You Need to Know About UGT1A1 Genetic Testing for Patients Treated With Irinotecan: A Practitioner-Friendly Guide. JCO Oncol. Pract. 2022, 18, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Pfizer: CAMPTOSAR® (Irinotecan HCl) Prescribing Information. 2014. Available online: https://labeling.pfizer.com/ShowLabeling.aspx?id=15268 (accessed on 28 September 2022).
- Sissung, T.M.; Barbier, R.H.; Price, D.K.; Plona, T.M.; Pike, K.M.; Mellott, S.D.; Baugher, R.N.; Whiteley, G.R.; Soppet, D.R.; Venzon, D.; et al. Comparison of Eight Technologies to Determine Genotype at the UGT1A1 (TA)(n) Repeat Polymorphism: Potential Clinical Consequences of Genotyping Errors? Int. J. Mol. Sci. 2020, 21, 896. [Google Scholar] [CrossRef] [PubMed]
- Hulshof, E.C.; de With, M.; de Man, F.M.; Creemers, G.-J.; Deiman, B.A.L.M.; Swen, J.J.; Houterman, S.; Koolen, S.L.W.; Bins, S.; Thijs, A.M.J.; et al. UGT1A1 genotype-guided dosing of irinotecan: A prospective safety and cost analysis in poor metaboliser patients. Eur. J. Cancer 2022, 162, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Hrafnkelsson, B.; Helgason, A.; Jonsson, G.F.; Gudbjartsson, D.F.; Jonsson, T.; Thorvaldsson, S.; Stefansson, H.; Steinthorsdottir, V.; Vidarsdottir, N.; Middleton, D.; et al. Evaluating differences in linkage disequilibrium between populations. Ann. Hum. Genet. 2010, 74, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Lanillos, J.; Carcajona, M.; Maietta, P.; Alvarez, S.; Rodriguez-Antona, C. Clinical pharmacogenetic analysis in 5,001 individuals with diagnostic Exome Sequencing data. NPJ Genom. Med. 2022, 7, 12. [Google Scholar] [CrossRef] [PubMed]
| rs Number | Coordinates | Alleles | MAF 1 | Distance | D’ | R2 | Correlated Alleles |
|---|---|---|---|---|---|---|---|
| rs34983651 | Chr2:234668879 | (-/AT) | 0.3253 | 0 | 1.0 | 1.0 | -=-,AT=AT |
| rs887829 | Chr2:234668570 | (C/T) | 354 | −309 | 0.9924 | 0.8663 | -=C,AT=T |
| rs111741722 | Chr2:234665983 | (A/G) | 0.3536 | −2896 | 0.9896 | 0.8628 | -=A,AT=G |
| rs4148325 | Chr2:234673309 | (C/T) | 0.3538 | 4430 | 0.9876 | 0.8588 | -=C,AT=T |
| rs35754645 | Chr2:234664586 | (TC/-) | 0.3524 | −4293 | 0.9839 | 0.8575 | -=TC,AT=- |
| rs6742078 | Chr2:234672639 | (G/T) | 0.3476 | 3760 | 0.9652 | 0.8428 | -=G,AT=T |
| rs4148324 | Chr2:234672722 | (T/G) | 0.3528 | 3843 | 0.9659 | 0.8249 | -=T,AT=G |
| rs3771341 | Chr2:234673239 | (G/A) | 0.3299 | 4360 | 0.8928 | 0.7807 | -=G,AT=A |
| rs10929302 | Chr2:234665782 | (G/A) | 0.3021 | −3097 | 0.9187 | 0.7579 | -=G,AT=A |
| rs6714634 | Chr2:234664765 | (T/C) | 0.3021 | −4114 | 0.9167 | 0.7547 | -=T,AT=C |
| rs34352510 | Chr2:234650562 | (T/C) | 0.3303 | −18317 | 0.8781 | 0.7538 | -=T,AT=C |
| R2 | D’ | Chi-sp | p Value | Population |
|---|---|---|---|---|
| 0.909558 | 1.0 | 180.0925 | <0.0001 | Utah residents from north and west Europe (CEU) (n = 198) |
| 0.9772 | 1.0 | 209.125 | <0.0001 | Iberian (n = 214) |
| 0.9767 | 1.0 | 209.0243 | <0.0001 | Toscani (n = 214) |
| 0.8932 | 1.0 | 176.8601 | <0.0001 | Finnish (n = 198) |
| 0.9719 | 1.0 | 179.8883 | <0.0001 | Britain (n = 182) |
| 0.9437 | 0.9855 | 949.3507 | <0.0001 | European (CEU, Iberian, Toscani, Finnish, Britain) (n = 1006) |
| 0.7646 | 1.0 | 1010.8077 | <0.0001 | African (n = 1322) |
| 0.9166 | 0.9936 | 636.152 | <0.0001 | Mixed American (Mexican, Puerto Ricans, Colombians, Peruvians) (n = 694) |
| 0.9565 | 0.9823 | 964.1246 | <0.0001 | East China (n = 1008) |
| 0.8404 | 0.9909 | 821.8782 | <0.0001 | South Asia (n = 978) |
| 0.8663 | 0.0024 | 4338.4445 | <0.0001 | All (n = 5008) |
| Homozygous A(TA)6TAA | Heterozygous A(TA)6TAA/A(TA)7TAA | Homozygous A(TA)7TAA | |
|---|---|---|---|
| PPV | 99.68% | 100% | 100% |
| NPV | 100% | 99.75% | 100% |
| Sensitivity | 100% | 99.67% | 100% |
| Specificity | 99.74% | 100% | 100% |
| Concept | Cost (EUR) 1 Sample per Run | Cost (EUR) 5 Samples per Run | Cost (EUR) 10 Samples per Run |
|---|---|---|---|
| Total cost | |||
| Fragment 1 | 41.8 | 20.8 | 17.3 |
| TaqMan 2 | 16.7 | 5.6 | 4.2 |
| Commercial 3 | 96.6 | 52.5 | 46.8 |
| Ratio Frag/TaqMan | 2.5 | 3.7 | 4.1 |
| Ratio Commercial/TaqMan | 5.8 | 9.4 | 11.1 |
| Reagents cost | |||
| Fragment 1 | 20.0 | 14.1 | 13.2 |
| TaqMan 2 | 4.1 | 1.5 | 1.2 |
| Commercial 3 | 73.4 | 44.9 | 41.3 |
| Ratio Frag/TaqMan | 4.8 | 9.8 | 11.4 |
| Ratio Commercial/TaqMan | 17.8 | 30.1 | 35.6 |
| Equipment | |||
| Fragment 1 | 0.6 | 0.6 | 0.6 |
| TaqMan 2 | 2.0 | 2.0 | 2.0 |
| Commercial 3 | 2.0 | 2.0 | 2.0 |
| Ratio Frag/TaqMan | 0.3 | 0.3 | 0.3 |
| Ratio Commercial/TaqMan | 1.0 | 1.0 | 1.0 |
| Staff | |||
| Fragment 1 | 21.2 | 5.6 | 3.5 |
| TaqMan 2 | 10.6 | 2.1 | 1.0 |
| Commercial 3 | 21.2 | 5.6 | 3.5 |
| Ratio Frag/TaqMan | 2.0 | 2.7 | 3.4 |
| Ratio Commercial/TaqMan | 1.0 | 1.0 | 1.0 |
| Task | Time per Sample | Time per 5 Samples | Time per 10 Samples |
|---|---|---|---|
| PCR and electrophoresis 1 | |||
| DNA extraction | 42′ | 45′ | 50′ |
| PCR | |||
| Purification | 20′ | 25′ | 30′ |
| Electrophoresis | 1 day | 1 day | 1 day |
| Analysis of results | 10′ | 15′ | 20′ |
| TaqMan probe 2 | |||
| DNA extraction | 5′ | 7′ | 10′ |
| Real-Time PCR | 35′ | 40′ | 45′ |
| Analysis of results | 5′ | 10′ | 15′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bravo-Gómez, A.; Salvador-Martín, S.; Zapata-Cobo, P.; Sanjurjo-Sáez, M.; López-Fernández, L.A. Genotyping of UGT1A1*80 as an Alternative to UGT1A1*28 Genotyping in Spain. Pharmaceutics 2022, 14, 2082. https://doi.org/10.3390/pharmaceutics14102082
Bravo-Gómez A, Salvador-Martín S, Zapata-Cobo P, Sanjurjo-Sáez M, López-Fernández LA. Genotyping of UGT1A1*80 as an Alternative to UGT1A1*28 Genotyping in Spain. Pharmaceutics. 2022; 14(10):2082. https://doi.org/10.3390/pharmaceutics14102082
Chicago/Turabian StyleBravo-Gómez, Adrián, Sara Salvador-Martín, Paula Zapata-Cobo, María Sanjurjo-Sáez, and Luis Andrés López-Fernández. 2022. "Genotyping of UGT1A1*80 as an Alternative to UGT1A1*28 Genotyping in Spain" Pharmaceutics 14, no. 10: 2082. https://doi.org/10.3390/pharmaceutics14102082
APA StyleBravo-Gómez, A., Salvador-Martín, S., Zapata-Cobo, P., Sanjurjo-Sáez, M., & López-Fernández, L. A. (2022). Genotyping of UGT1A1*80 as an Alternative to UGT1A1*28 Genotyping in Spain. Pharmaceutics, 14(10), 2082. https://doi.org/10.3390/pharmaceutics14102082