Mass Cytometry Exploration of Immunomodulatory Responses of Human Immune Cells Exposed to Silver Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Silver Nanoparticles
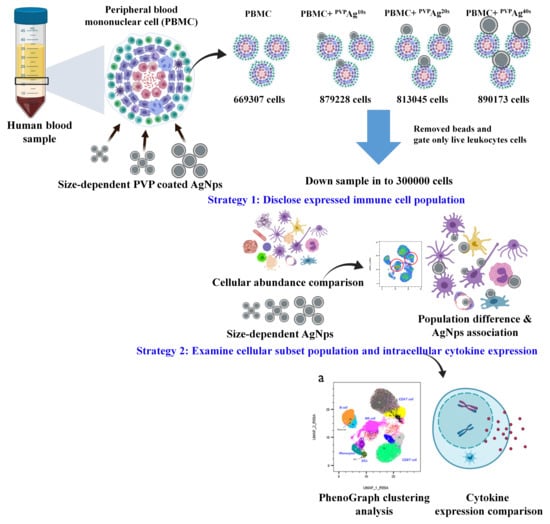
2.2. Isolation of PBMCs from Whole Blood
2.3. Stimulation of PBMCs and Exposure to Ag NPs
2.4. Surface and Intracellular Marker Staining
2.5. Mass Cytometer Setup, Calibration, and Data Acquisition
2.6. Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Physicochemical Characteristics of Silver Nanoparticles
3.2. Cellular Association of Ag NPs on hPBMCs: Effects of Cell Type and NP Core Size
3.3. Variations of Immune Cell Populations: Effect of the Core Sizes of Ag NPs
3.4. Variations of PhenoGraph Subclusters of Immune Cell Populations: Effect of the Core Sizes of Ag NPs
3.5. Variations of in Intracellular Cytokine Expression: Effect of the Core Size of Ag NPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flores, A.M.; Ye, J.; Jarr, K.-U.; Hosseini-Nassab, N.; Smith, B.R.; Leeper, N.J. Nanoparticle Therapy for Vascular Diseases. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Kreuter, J.; Gelperina, S. Use of Nanoparticles for Cerebral Cancer. Tumori J. 2008, 94, 271–277. [Google Scholar] [CrossRef]
- Johnston, B.D.; Scown, T.M.; Moger, J.; Cumberland, S.A.; Baalousha, M.; Linge, K.; van Aerle, R.; Jarvis, K.; Lead, J.R.; Tyler, C.R. Bioavailability of Nanoscale Metal Oxides TiO2, CeO2, and ZnO to Fish. Environ. Sci. Technol. 2010, 44, 1144–1151. [Google Scholar] [CrossRef] [Green Version]
- Vance, M.E.; Kuiken, T.; Vejerano, E.P.; McGinnis, S.P.; Hochella, M.F.; Rejeski, D.; Hull, M.S. Nanotechnology in the real world: Redeveloping the nanomaterial consumer products inventory. Beilstein J. Nanotechnol. 2015, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Baker, C.; Pradhan, A.; Pakstis, L.; Pochan Darrin, J.; Shah, S.I. Synthesis and Antibacterial Properties of Silver Nanoparticles. J. Nanosci. Nanotechnol. 2005, 5, 244–249. [Google Scholar] [CrossRef]
- Samberg, M.E.; Orndorff, P.E.; Monteiro-Riviere, N.A. Antibacterial efficacy of silver nanoparticles of different sizes, surface conditions and synthesis methods. Nanotoxicology 2011, 5, 244–253. [Google Scholar] [CrossRef]
- Elsabahy, M.; Wooley, K.L. Cytokines as biomarkers of nanoparticle immunotoxicity. Chem. Soc. Rev. 2013, 42, 5552–5576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greulich, C.; Diendorf, J.; Simon, T.; Eggeler, G.; Epple, M.; Köller, M. Uptake and intracellular distribution of silver nanoparticles in human mesenchymal stem cells. Acta Biomater. 2011, 7, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Jun, L.; Qiuzhen, W.; Qingguo, M. The effects of project uncertainty and risk management on IS development project performance: A vendor perspective. Int. J. Proj. Manag. 2011, 29, 923–933. [Google Scholar] [CrossRef]
- Bian, Y.; Kim, K.; Ngo, T.; Kim, I.; Bae, O.N.; Lim, K.M.; Chung, J.H. Silver nanoparticles promote procoagulant activity of red blood cells: A potential risk of thrombosis in susceptible population. Part. Fibre Toxicol. 2019, 16, 9. [Google Scholar] [CrossRef]
- AshaRani, P.V.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and Genotoxicity of Silver Nanoparticles in Human Cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef]
- Ghosh, M.; Manivannan, J.; Sinha, S.; Chakraborty, A.; Mallick, S.K.; Bandyopadhyay, M.; Mukherjee, A. In vitro and in vivo genotoxicity of silver nanoparticles. Mutat. Res. 2012, 749, 60–69. [Google Scholar] [CrossRef]
- Martinez-Gutierrez, F.; Thi, E.P.; Silverman, J.M.; de Oliveira, C.C.; Svensson, S.L.; Vanden Hoek, A.; Sánchez, E.M.; Reiner, N.E.; Gaynor, E.C.; Pryzdial, E.L.G.; et al. Antibacterial activity, inflammatory response, coagulation and cytotoxicity effects of silver nanoparticles. Nanomedicine 2012, 8, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Ivask, A.; Kurvet, I.; Kasemets, K.; Blinova, I.; Aruoja, V.; Suppi, S.; Vija, H.; Käkinen, A.; Titma, T.; Heinlaan, M.; et al. Size-dependent toxicity of silver nanoparticles to bacteria, yeast, algae, crustaceans and mammalian cells in vitro. PLoS ONE 2014, 9, e102108. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Gödel, P.; Subklewe, M.; Stemmler, H.J.; Schlößer, H.A.; Schlaak, M.; Kochanek, M.; Böll, B.; von Bergwelt-Baildon, M.S. Cytokine release syndrome. J. Immunother. Cancer 2018, 6, 56. [Google Scholar] [CrossRef] [Green Version]
- Paradiso, A.; Domingo, G.C.; Blanco, E.; Buscaglia, A.; Fortunato, S.; Marsoni, M.; Scarcia, P.; Caretto, S.; Vannini, C.; de Pinto, M.C. Cyclic AMP mediates heat stress response by the control of redox homeostasis and ubiquitin-proteasome system. Plant Cell Environ. 2020, 43, 2727–2742. [Google Scholar] [CrossRef] [PubMed]
- Alijagic, A.; Gaglio, D.; Napodano, E.; Russo, R.; Costa, C.; Benada, O.; Kofroňová, O.; Pinsino, A. Titanium dioxide nanoparticles temporarily influence the sea urchin immunological state suppressing inflammatory-relate gene transcription and boosting antioxidant metabolic activity. J. Hazard. Mater. 2020, 384, 121389. [Google Scholar] [CrossRef] [PubMed]
- Boraschi, D.; Italiani, P.; Palomba, R.; Decuzzi, P.; Duschl, A.; Fadeel, B.; Moghimi, S.M. Nanoparticles and innate immunity: New perspectives on host defence. Semin. Immunol. 2017, 34, 33–51. [Google Scholar] [CrossRef]
- Auguste, M.; Lasa, A.; Pallavicini, A.; Gualdi, S.; Vezzulli, L.; Canesi, L. Exposure to TiO2 nanoparticles induces shifts in the microbiota composition of Mytilus galloprovincialis hemolymph. Sci. Total Environ. 2019, 670, 129–137. [Google Scholar] [CrossRef]
- Hawkins, S.J.; Crompton, L.A.; Sood, A.; Saunders, M.; Boyle, N.T.; Buckley, A.; Minogue, A.M.; McComish, S.F.; Jiménez-Moreno, N.; Cordero-Llana, O.; et al. Nanoparticle-induced neuronal toxicity across placental barriers is mediated by autophagy and dependent on astrocytes. Nat. Nanotechnol. 2018, 13, 427–433. [Google Scholar] [CrossRef]
- Savage, D.T.; Hilt, J.Z.; Dziubla, T.D. In Vitro Methods for Assessing Nanoparticle Toxicity. Methods Mol. Biol. 2019, 1894, 1–29. [Google Scholar] [CrossRef]
- Fraser, J.A.; Kemp, S.; Young, L.; Ross, M.; Prach, M.; Hutchison, G.R.; Malone, E. Silver nanoparticles promote the emergence of heterogeneic human neutrophil sub-populations. Sci. Rep. 2018, 8, 7506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panzarini, E.; Mariano, S.; Vergallo, C.; Fimia, G.M.; Dini, L.; Mura, F.; Rossi, M.; Serra, A.; Casciaro, S. Glucose capped silver nanoparticles enter HeLa cells and induce S and G2/M arrest. In Proceedings of the 2015 1st Workshop on Nanotechnology in Instrumentation and Measurement (NANOFIM), Lecce, Italy, 24–25 July 2015; pp. 72–76. [Google Scholar]
- Li, L.; Fernández-Cruz, M.L.; Connolly, M.; Conde, E.; Fernández, M.; Schuster, M.; Navas, J.M. The potentiation effect makes the difference: Non-toxic concentrations of ZnO nanoparticles enhance Cu nanoparticle toxicity in vitro. Sci. Total Environ. 2015, 505, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Henriksen-Lacey, M.; Carregal-Romero, S.; Liz-Marzán, L.M. Current Challenges toward In Vitro Cellular Validation of Inorganic Nanoparticles. Bioconjug. Chem. 2017, 28, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Kroll, A.; Pillukat, M.H.; Hahn, D.; Schnekenburger, J. Current in vitro methods in nanoparticle risk assessment: Limitations and challenges. Eur. J. Pharm. Biopharm. 2009, 72, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Monteiro-Riviere, N.A.; Inman, A.O.; Zhang, L.W. Limitations and relative utility of screening assays to assess engineered nanoparticle toxicity in a human cell line. Toxicol. Appl. Pharmacol. 2009, 234, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Bandura, D.R.; Baranov, V.I.; Ornatsky, O.I.; Antonov, A.; Kinach, R.; Lou, X.; Pavlov, S.; Vorobiev, S.; Dick, J.E.; Tanner, S.D. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009, 81, 6813–6822. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2020, arXiv:1802.03426. [Google Scholar]
- Levine, J.H.; Simonds, E.F.; Bendall, S.C.; Davis, K.L.; Amir, E.D.; Tadmor, M.D.; Litvin, O.; Fienberg, H.G.; Jager, A.; Zunder, E.R.; et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 2015, 162, 184–197. [Google Scholar] [CrossRef] [Green Version]
- Fienberg, H.G.; Simonds, E.F.; Fantl, W.J.; Nolan, G.P.; Bodenmiller, B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytom. A 2012, 81, 467–475. [Google Scholar] [CrossRef]
- Ha, M.K.; Kwon, S.J.; Choi, J.S.; Nguyen, N.T.; Song, J.; Lee, Y.; Kim, Y.E.; Shin, I.; Nam, J.W.; Yoon, T.H. Mass Cytometry and Single-Cell RNA-seq Profiling of the Heterogeneity in Human Peripheral Blood Mononuclear Cells Interacting with Silver Nanoparticles. Small 2020, 16, e1907674. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.K.; Choi, J.-S.; Kwon, S.J.; Song, J.; Lee, Y.; Kim, Y.-E.; Yoon, T.H. Mass cytometric study on the heterogeneity in cellular association and cytotoxicity of silver nanoparticles in primary human immune cells. Environ. Sci. Nano 2020, 7, 1102–1114. [Google Scholar] [CrossRef]
- Gerner, M.Y.; Torabi-Parizi, P.; Germain, R.N. Strategically Localized Dendritic Cells Promote Rapid T Cell Responses to Lymph-Borne Particulate Antigens. Immunity 2015, 42, 172–185. [Google Scholar] [CrossRef] [Green Version]
- Curtsinger, J.M.; Schmidt, C.S.; Mondino, A.; Lins, D.C.; Kedl, R.M.; Jenkins, M.K.; Mescher, M.F. Inflammatory Cytokines Provide a Third Signal for Activation of Naive CD4+ and CD8+ T Cells. J. Immunol. 1999, 162, 3256–3262. [Google Scholar]
- Ramachandran, G.; Kumar, A.K.H.; Kannan, T.; Bhavani, P.K.; Kumar, S.R.; Gangadevi, N.P.; Banurekha, V.; Sudha, V.; Venkatesh, S.; Ravichandran, N.; et al. Low Serum Concentrations of Rifampicin and Pyrazinamide Associated with Poor Treatment Outcomes in Children with Tuberculosis Related to HIV Status. Pediatr. Infect. Dis. J. 2016, 35, 530–534. [Google Scholar] [CrossRef]
- Ha, M.K.; Shim, Y.J.; Yoon, T.H. Effects of agglomeration on in vitro dosimetry and cellular association of silver nanoparticles. Environ. Sci. Nano 2018, 5, 446–455. [Google Scholar] [CrossRef]
- Minghetti, M.; Schirmer, K. Effect of media composition on bioavailability and toxicity of silver and silver nanoparticles in fish intestinal cells (RTgutGC). Nanotoxicology 2016, 10, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Walczyk, D.; Bombelli, F.B.; Monopoli, M.P.; Lynch, I.; Dawson, K.A. What the Cell “Sees” in Bionanoscience. J. Am. Chem. Soc. 2010, 132, 5761–5768. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical−Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef]
- Ellis, L.-J.A.; Baalousha, M.; Valsami-Jones, E.; Lead, J.R. Seasonal variability of natural water chemistry affects the fate and behaviour of silver nanoparticles. Chemosphere 2018, 191, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.-J.A.; Lynch, I. Mechanistic insights into toxicity pathways induced by nanomaterials in Daphnia magna from analysis of the composition of the acquired protein corona. Environ. Sci. Nano 2020, 7, 3343–3359. [Google Scholar] [CrossRef]
- Ostermeyer, A.-K.; Kostigen Mumuper, C.; Semprini, L.; Radniecki, T. Influence of Bovine Serum Albumin and Alginate on Silver Nanoparticle Dissolution and Toxicity to Nitrosomonas europaea. Environ. Sci. Technol. 2013, 47, 14403–14410. [Google Scholar] [CrossRef] [PubMed]
- Levak, M.; Burić, P.; Dutour Sikirić, M.; Domazet Jurašin, D.; Mikac, N.; Bačić, N.; Drexel, R.; Meier, F.; Jakšić, Ž.; Lyons, D.M. Effect of Protein Corona on Silver Nanoparticle Stabilization and Ion Release Kinetics in Artificial Seawater. Environ. Sci. Technol. 2017, 51, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Peretyazhko, T.S.; Zhang, Q.; Colvin, V.L. Size-Controlled Dissolution of Silver Nanoparticles at Neutral and Acidic pH Conditions: Kinetics and Size Changes. Environ. Sci. Technol. 2014, 48, 11954–11961. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Kim, E.; Cho, H.-J.; Kang, T.; Kim, B.; Kim, M.Y.; Kim, Y.S.; Song, N.W.; Lee, J.-S.; Jeong, J. The Relationship between Dissolution Behavior and the Toxicity of Silver Nanoparticles on Zebrafish Embryos in Different Ionic Environments. Nanomaterials 2018, 8, 652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groh, K.J.; Dalkvist, T.; Piccapietra, F.; Behra, R.; Suter, M.J.-F.; Schirmer, K. Critical influence of chloride ions on silver ion-mediated acute toxicity of silver nanoparticles to zebrafish embryos. Nanotoxicology 2015, 9, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Arabeyyat, Z.H.; Xin, Q.; Paunov, V.N.; Dale, I.J.F.; Lloyd Mills, R.I.; Rotchell, J.M.; Cheng, J. Silver Nanoparticles in Zebrafish (Danio rerio) Embryos: Uptake, Growth and Molecular Responses. Int. J. Mol. Sci. 2020, 21, 1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubbell, J.A.; Thomas, S.N.; Swartz, M.A. Materials engineering for immunomodulation. Nature 2009, 462, 449–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kettler, K.; Veltman, K.; van de Meent, D.; van Wezel, A.; Hendriks, A.J. Cellular uptake of nanoparticles as determined by particle properties, experimental conditions, and cell type. Environ. Toxicol. Chem. 2014, 33, 481–492. [Google Scholar] [CrossRef]
- Buono, C.; Anzinger, J.J.; Amar, M.; Kruth, H.S. Fluorescent pegylated nanoparticles demonstrate fluid-phase pinocytosis by macrophages in mouse atherosclerotic lesions. J. Clin. Investig. 2009, 119, 1373–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Xu, H.; Han, Y.; Dai, W.; Hao, W.; Wang, C.; Gu, N.; Xu, H.; Cao, J. The internalization pathway, metabolic fate and biological effect of superparamagnetic iron oxide nanoparticles in the macrophage-like RAW264.7 cell. Sci. China Life Sci. 2011, 54, 793–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pricop, D. Maria Andrieş Endocytosis and Exocytosis of Gold Nanoparticles. Rom. J. Biophys. 2015, 25, 63–71. [Google Scholar]
- Kuhn, D.A.; Vanhecke, D.; Michen, B.; Blank, F.; Gehr, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Different endocytotic uptake mechanisms for nanoparticles in epithelial cells and macrophages. Beilstein J. Nanotechnol. 2014, 5, 1625–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cifuentes-Rius, A.; Desai, A.; Yuen, D.; Johnston, A.P.R.; Voelcker, N.H. Inducing immune tolerance with dendritic cell-targeting nanomedicines. Nat. Nanotechnol. 2021, 16, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Tomić, S.; Đokić, J.; Vasilijić, S.; Ogrinc, N.; Rudolf, R.; Pelicon, P.; Vučević, D.; Milosavljević, P.; Janković, S.; Anžel, I.; et al. Size-Dependent Effects of Gold Nanoparticles Uptake on Maturation and Antitumor Functions of Human Dendritic Cells In Vitro. PLoS ONE 2014, 9, e96584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zolnik, B.S.; González-Fernández, A.; Sadrieh, N.; Dobrovolskaia, M.A. Nanoparticles and the immune system. Endocrinology 2010, 151, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.-H.; Chang, L.W.; Lin, P. Metal-Based Nanoparticles and the Immune System: Activation, Inflammation, and Potential Applications. BioMed Res. Int. 2015, 2015, 143720. [Google Scholar] [CrossRef] [PubMed]
- Turvey, S.E.; Broide, D.H. Innate immunity. J. Allergy Clin. Immunol. 2010, 125, S24–S32. [Google Scholar] [CrossRef] [PubMed]
- ten Broeke, T.; Wubbolts, R.; Stoorvogel, W. MHC class II antigen presentation by dendritic cells regulated through endosomal sorting. Cold Spring Harb. Perspect. Biol. 2013, 5, a016873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochweller, K.; Wabnitz, G.H.; Samstag, Y.; Suffner, J.; Hämmerling, G.J.; Garbi, N. Dendritic cells control T cell tonic signaling required for responsiveness to foreign antigen. Proc. Natl. Acad. Sci. USA 2010, 107, 5931–5936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanier, L.L. Nk Cell Recognition. Annu. Rev. Immunol. 2004, 23, 225–274. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Malissen, B. Innate and adaptive immunity: Specificities and signaling hierarchies revisited. Nat. Immunol. 2005, 6, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jaw, J.J.; Stutzman, N.C.; Zou, Z.; Sun, P.D. Natural killer cell-produced IFN-γ and TNF-α induce target cell cytolysis through up-regulation of ICAM-1. J. Leukoc. Biol. 2012, 91, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Roda, J.M.; Parihar, R.; Magro, C.; Nuovo, G.J.; Tridandapani, S.; Carson, W.E. Natural Killer Cells Produce T Cell–Recruiting Chemokines in Response to Antibody-Coated Tumor Cells. Cancer Res. 2006, 66, 517–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauriat, C.; Long, E.O.; Ljunggren, H.-G.; Bryceson, Y.T. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 2010, 115, 2167–2176. [Google Scholar] [CrossRef] [Green Version]
- Swain, S.L.; McKinstry, K.K.; Strutt, T.M. Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 2012, 12, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Schuemann, J.; Bagley, A.F.; Berbeco, R.; Bromma, K.; Butterworth, K.T.; Byrne, H.L.; Chithrani, B.D.; Cho, S.H.; Cook, J.R.; Favaudon, V.; et al. Roadmap for metal nanoparticles in radiation therapy: Current status, translational challenges, and future directions. Phys. Med. Biol. 2020, 65, 21RM02. [Google Scholar] [CrossRef] [PubMed]
- Penninckx, S.; Heuskin, A.-C.; Michiels, C.; Lucas, S. Gold Nanoparticles as a Potent Radiosensitizer: A Transdisciplinary Approach from Physics to Patient. Cancers 2020, 12, 2021. [Google Scholar] [CrossRef] [PubMed]
- Penninckx, S.; Heuskin, A.-C.; Michiels, C.; Lucas, S. The role of thioredoxin reductase in gold nanoparticle radiosensitization effects. Nanomedicine 2018, 13, 2917–2937. [Google Scholar] [CrossRef] [PubMed]







| Cell Type | Phenograph Cluster | Marker Expression |
|---|---|---|
| Naïve CD4+ T cell | #2 | CD3+CD4+CD27+CD38+CD45RA+ |
| #3 | CD3+CD4+CD27+CD38+CD45RA+ | |
| #5 | CD3+CD4+CD27+CD38+CD45RA+ | |
| #9 | CD3+CD4+CD27+CD38+CD45RA+ | |
| Effector CD4+ T cell | #18 | CD3+CD4+CD27-CD38-CD45RA- |
| Memory CD4+ T cell | #6 | CD3+CD4+CD27+CD38+CD45RA- |
| #10 | CD3+CD4+CD27+CD38-CD45RA-HLA-DRmid | |
| Naïve CD8+ T cell | #1 | CD3+CD8+CD27+CD38+CD45RA+ |
| #8 | CD3+CD8+CD27+CD38+CD45RA+ | |
| #11 | CD3+CD8+CD27+CD38-CD45RA+ | |
| #19 | CD3+CD8+CD27+CD38midCD45RA+ | |
| #21 | CD3+CD8+CD27+CD38midCD45RA+ | |
| Effector CD8+ T cell | #14 | CD3+CD8+CD27+CD38-CD45RA+ |
| Memory CD8+ T cell | #15 | CD3+CD8+CD27+CD38midCD45RA-HLA-DRmid |
| Naïve B cell | #7 | CD19+CD20+CD27-CD38+CD45RA+HLA-DR+ |
| Memory B cell | #13 | CD19+CD20+CD27+CD38+CD45RA+HLA-DR+CD16midCD11c+ |
| NK cell | #4 | CD19-CD8a+CD27-CD38+CD45RA+CD11c+ |
| #17 | CD19-CD8a+CD27midCD38+CD45RA+ | |
| #22 | CD19-CD8a-CD27-CD38+CD45RA-HLA-DR+CD11c+CD123+ | |
| #24 | CD19-CD8a-CD27-CD38+CD45RA+HLA-DR+CD123+ | |
| Classical monocyte | #16 | CD14+CD11c+CD123midHLA-DR+CD38+ |
| Nonclassical monocyte | #20 | CD14-CD11c+CD123midHLA-DR+CD38-CD66a+ |
| Dendritic cell | #23 | CD11c+CD123+HLA-DR+CD38+ |
| Unassigned | #12 | CD3+CD4+CD27+CD38-CD45RA+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, J.; Ha, M.; Perumalsamy, H.; Lee, Y.; Song, J.; Yoon, T.-H. Mass Cytometry Exploration of Immunomodulatory Responses of Human Immune Cells Exposed to Silver Nanoparticles. Pharmaceutics 2022, 14, 630. https://doi.org/10.3390/pharmaceutics14030630
Bae J, Ha M, Perumalsamy H, Lee Y, Song J, Yoon T-H. Mass Cytometry Exploration of Immunomodulatory Responses of Human Immune Cells Exposed to Silver Nanoparticles. Pharmaceutics. 2022; 14(3):630. https://doi.org/10.3390/pharmaceutics14030630
Chicago/Turabian StyleBae, Jiwon, My Ha, Haribalan Perumalsamy, Yangsoon Lee, Jaewoo Song, and Tae-Hyun Yoon. 2022. "Mass Cytometry Exploration of Immunomodulatory Responses of Human Immune Cells Exposed to Silver Nanoparticles" Pharmaceutics 14, no. 3: 630. https://doi.org/10.3390/pharmaceutics14030630
APA StyleBae, J., Ha, M., Perumalsamy, H., Lee, Y., Song, J., & Yoon, T. -H. (2022). Mass Cytometry Exploration of Immunomodulatory Responses of Human Immune Cells Exposed to Silver Nanoparticles. Pharmaceutics, 14(3), 630. https://doi.org/10.3390/pharmaceutics14030630