Plasma-Enabled Smart Nanoexosome Platform as Emerging Immunopathogenesis for Clinical Viral Infection
Abstract
:1. Introduction
2. Background of Smart Nanoexosomes
2.1. The Biogenesis of Nanoexosomes
2.2. Roles of Smart Nanoexosomes of RNA Viruses
2.3. DNA Viruses
| Virus | Genome | Features and Response | Ref. |
|---|---|---|---|
| HIV-1 | ssRNA positive sense | Broad host range (non-dividing cells) Long-term, inducible expression | [92,93,94,95] |
| HIV-2 | Chromosomal integration | ||
| NDV | ssRNA negative sense | Replication in tumor cells Improved oncolytic vectors | [96,97,98] |
| HPV | dsDNA, papillomavirus | Double-stranded | [99,100,101] |
| B19V | ssDNA, parvovirus | dispensable for cell cycle arrest at phase G2/M | [102,103,104] |
| Herpesvirus | dsDNA | Risk of recombination with latently herpes simplex virus- infected cells | [105] |
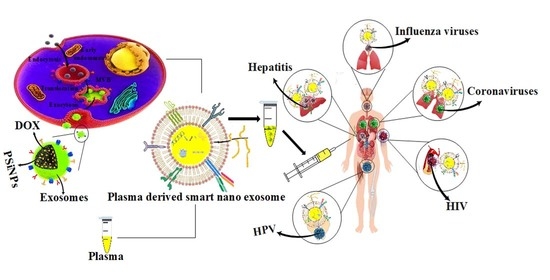
3. Plasma Derived Smart Nanoexosomes
4. Clinical Translation Pathway
4.1. Coronaviruses
4.2. Influenza Viruses
4.3. HPV
4.4. Hepatitis and HIV
5. Smart Nanoexosomes as Biosensors
6. Challenges with Smart Nanoexosome Therapeutics
7. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Verrilli, M.A.; Court, F.A. Exosomes: Mediators of communication in eukaryotes. Biol. Res. 2013, 46, 5–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Biomark. Med. 2013, 7, 769–778. [Google Scholar] [CrossRef]
- Fernandes, M.; Lopes, I.; Teixeira, J.; Botelho, C.; Gomes, A. Exosome-like Nanoparticles: A New Type of Nanocarrier. Curr. Med. Chem. 2020, 27, 3888–3905. [Google Scholar] [CrossRef]
- Gholami, A.; Hashemi, S.A.; Yousefi, K.; Mousavi, S.M.; Chiang, W.-H.; Ramakrishna, S.; Mazraedoost, S.; Alizadeh, A.; Omidifar, N.; Behbudi, G.; et al. 3D Nanostructures for Tissue Engineering, Cancer Therapy, and Gene Delivery. J. Nanomater. 2020, 2020, 1–24. [Google Scholar] [CrossRef]
- Ludwig, A.-K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012, 44, 11–15. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Zarei, M.; Hashemi, S.A.R. Polydopamine for Biomedical Application and Drug Delivery System. Med. Chem. 2018, 8, 218–229. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [Green Version]
- Kazemi, K.; Ghahramani, Y.; Kalashgrani, M.Y. Nano biofilms: An emerging biotechnology applications. Adv. Appl. NanoBio-Technol. 2022, 3, 8–15. [Google Scholar]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousavi, S.M.; Low, F.W.; Hashemi, S.A.; Lai, C.W.; Ghasemi, Y.; Soroshnia, S.; Savardashtaki, A.; Babapoor, A.; Rumjit, N.P.; Goh, S.M.; et al. Development of graphene based nanocomposites towards medical and biological applications. Artif. Cells Nanomed. Biotechnol. 2020, 48, 1189–1205. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoorvogel, W.; Kleijmeer, M.J.; Geuze, H.J.; Raposo, G. The Biogenesis and Functions of Exosomes. Traffic 2002, 3, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Alenquer, M.; Amorim, M.J. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef] [Green Version]
- Bahrani, S.; Hashemi, S.A.; Mousavi, S.M.; Azhdari, R. Zinc-based metal–organic frameworks as nontoxic and biodegradable platforms for biomedical applications: Review study. Drug Metab. Rev. 2019, 51, 356–377. [Google Scholar] [CrossRef]
- Petrik, J. Immunomodulatory effects of exosomes produced by virus-infected cells. Transfus. Apher. Sci. 2016, 55, 84–91. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [Green Version]
- Stuffers, S.; Wegner, C.S.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavík, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greening, D.W.; Simpson, R.J. Understanding extracellular vesicle diversity–current status. Expert Rev. Proteom. 2018, 15, 887–910. [Google Scholar] [CrossRef]
- Latifkar, A.; Hur, Y.H.; Sanchez, J.C.; Cerione, R.A.; Antonyak, M.A. New insights into extracellular vesicle biogenesis and function. J. Cell Sci. 2019, 132, jcs222406. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Burrola, S.; Wu, J.; Ding, W.-Q. Extracellular Vesicles in the Development of Cancer Therapeutics. Int. J. Mol. Sci. 2020, 21, 6097. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Mousavi, S.; Hashemi, S.; Bahrani, S.; Yousefi, K.; Behbudi, G.; Babapoor, A.; Omidifar, N.; Lai, C.; Gholami, A.; Chiang, W.-H. Recent Advancements in Polythiophene-Based Materials and their Biomedical, Geno Sensor and DNA Detection. Int. J. Mol. Sci. 2021, 22, 6850. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2014, 1841, 108–120. [Google Scholar] [CrossRef]
- Kadiu, I.; Narayanasamy, P.; Dash, P.K.; Zhang, W.; Gendelman, H.E. Biochemical and Biologic Characterization of Exosomes and Microvesicles as Facilitators of HIV-1 Infection in Macrophages. J. Immunol. 2012, 189, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Wiley, R.D.; Gummuluru, S. Immature dendritic cell-derived exosomes can mediate HIV-1 trans infection. Proc. Natl. Acad. Sci. USA 2006, 103, 738–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenassi, M.; Cagney, G.; Liao, M.; Vaupotic, T.; Bartholomeeusen, K.; Cheng, Y.; Krogan, N.J.; Plemenitas, A.; Peterlin, B.M. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010, 11, 110–122. [Google Scholar] [CrossRef] [Green Version]
- Chahar, H.S.; Bao, X.; Casola, A. Exosomes and Their Role in the Life Cycle and Pathogenesis of RNA Viruses. Viruses 2015, 7, 3204–3225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aqil, M.; Naqvi, A.R.; Mallik, S.; Bandyopadhyay, S.; Maulik, U.; Jameel, S. The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. J. Extracell. Vesicles 2014, 3, 23129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaefer, M.R.; Wonderlich, E.R.; Roeth, J.F.; Leonard, J.A.; Collins, K.L. HIV-1 Nef Targets MHC-I and CD4 for Degradation Via a Final Common β-COP–Dependent Pathway in T Cells. PLoS Pathog. 2008, 4, e1000131. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Wu, N.; Gan, X.; Yan, W.; Morrell, J.C.; Gould, S.J. Higher-Order Oligomerization Targets Plasma Membrane Proteins and HIV Gag to Exosomes. PLoS Biol. 2007, 5, e158. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, S.A.; Mousavi, S.M.; Bahrani, S.; Ramakrishna, S.; Babapoor, A.; Chiang, W.-H. Coupled graphene oxide with hybrid metallic nanoparticles as potential electrochemical biosensors for precise detection of ascorbic acid within blood. Anal. Chim. Acta 2020, 1107, 183–192. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Pawliczek, T.; Crump, C.M. Herpes Simplex Virus Type 1 Production Requires a Functional ESCRT-III Complex but Is Independent of TSG101 and ALIX Expression. J. Virol. 2009, 83, 11254–11264. [Google Scholar] [CrossRef] [Green Version]
- Votteler, J.; Sundquist, W.I. Virus Budding and the ESCRT Pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, A.; Iordanskiy, S.; Das, R.; Van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes Derived from HIV-1-infected Cells Contain Trans-activation Response Element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousavi, S.M.; Zarei, M.; Hashemi, S.A.; Babapoor, A.; Amani, A.M. A conceptual review of rhodanine: Current applications of antiviral drugs, anticancer and antimicrobial activities. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1132–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousavi, S.M.; Soroshnia, S.; Hashemi, S.A.; Babapoor, A.; Ghasemi, Y.; Savardashtaki, A.; Amani, A.M. Graphene nano-ribbon based high potential and efficiency for DNA, cancer therapy and drug delivery applications. Drug Metab. Rev. 2019, 51, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Bernard, M.; Zhao, H.; Yue, S.C.; Anandaiah, A.; Koziel, H.; Tachado, S.D. Novel HIV-1 MiRNAs Stimulate TNFα Release in Human Macrophages via TLR8 Signaling Pathway. PLoS ONE 2014, 9, e106006. [Google Scholar] [CrossRef]
- Konadu, K.A.; Chu, J.; Huang, M.B.; Amancha, P.K.; Armstrong, W.S.; Powell, M.D.; Villinger, F.; Bond, V.C. Association of Cytokines With Exosomes in the Plasma of HIV-1–Seropositive Individuals. J. Infect. Dis. 2015, 211, 1712–1716. [Google Scholar] [CrossRef]
- Gholami, A.; Mousavi, S.M.; Hashemi, S.A.; Ghasemi, Y.; Chiang, W.-H.; Parvin, N. Current trends in chemical modifications of magnetic nanoparticles for targeted drug delivery in cancer chemotherapy. Drug Metab. Rev. 2020, 52, 205–224. [Google Scholar]
- Mack, M.; Kleinschmidt, A.; Brühl, H.; Klier, C.; Nelson, P.J.; Cihak, J.; Plachý, J.; Stangassinger, M.; Erfle, V.; Schlöndorff, D. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: A mechanism for cellular human immunodeficiency virus 1 infection. Nat. Med. 2000, 6, 769–775. [Google Scholar] [CrossRef]
- Haque, S.; Vaiselbuh, S. CD19 Chimeric Antigen Receptor-Exosome Targets CD19 Positive B-lineage Acute Lymphocytic Leukemia and Induces Cytotoxicity. Cancers 2021, 13, 1401. [Google Scholar] [CrossRef]
- Fang, W.H.; Agrawal, D.K.; Thankam, F.G. “Smart Exosomes”: A Smart Approach for Tendon Regeneration. Tissue Eng. Part B Rev. 2021. ahead of print. [Google Scholar] [CrossRef]
- Ji, P.; Yang, Z.; Li, H.; Wei, M.; Yang, G.; Xing, H.; Li, Q. Smart exosomes with lymph node homing and immune-amplifying capacities for enhanced immunotherapy of metastatic breast cancer. Mol. Ther. Nucleic Acids 2021, 26, 987–996. [Google Scholar] [CrossRef]
- Harishkumar, M.; Radha, M.; Yuichi, N.; Muthukalianan, G.K.; Kaoru, O.; Shiomori, K.; Sakai, K.; Nozomi, W. Designer Exosomes: Smart Nano-Communication Tools for Translational Medicine. Bioengineering 2021, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Hashemi, S.A.; Kalashgrani, M.Y.; Gholami, A.; Omidifar, N.; Babapoor, A.; Rao, N.V.; Chiang, W.-H. Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis. Biosensors 2022, 12, 286. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Zarei, M.; Bahrani, S.; Savardashtaki, A.; Esmaeili, H.; Lai, C.W.; Mazraedoost, S.; Abassi, M.; Ramavandi, B. Data on cytotoxic and antibacterial activity of synthesized Fe3O4 nanoparticles using Malva sylvestris. Data Brief. 2019, 28, 104929. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- White, I.J.; Bailey, L.M.; Aghakhani, M.R.; Moss, S.E.; Futter, C.E. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2005, 25, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulrahman, B.; Abdelaziz, D.H.; Schatzl, H.M. Autophagy regulates exosomal release of prions in neuronal cells. J. Biol. Chem. 2018, 293, 8956–8968. [Google Scholar] [CrossRef] [Green Version]
- Agromayor, M.; Soler, N.; Caballe, A.; Kueck, T.; Freund, S.M.; Allen, M.D.; Bycroft, M.; Perisic, O.; Ye, Y.; McDonald, B.; et al. The UBAP1 Subunit of ESCRT-I Interacts with Ubiquitin via a SOUBA Domain. Structure 2012, 20, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.A.; Huang, M.-B.; Campbell, P.E.; Roth, W.W.; Campbell, T.; Khan, M.; Newman, G.; Villinger, F.; Powell, M.D.; Bond, V.C. Genetic Characterization of HIV Type 1 Nef-Induced Vesicle Secretion. AIDS Res. Hum. Retrovir. 2010, 26, 173–192. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Pant, S.; Hilton, H.; Burczynski, M.E. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem. Pharmacol. 2012, 83, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- Rozmyslowicz, T.; Majka, M.; Kijowski, J.; Murphy, S.L.; Conover, D.O.; Poncz, M.; Ratajczak, J.; Gaulton, G.N.; Ratajczak, M.Z. Platelet- and megakaryocyte-derived microparticles transfer CXCR4 receptor to CXCR4-null cells and make them susceptible to infection by X4-HIV. AIDS 2003, 17, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Arenaccio, C.; Chiozzini, C.; Columba-Cabezas, S.; Manfredi, F.; Affabris, E.; Baur, A.; Federico, M. Exosomes from Human Immunodeficiency Virus Type 1 (HIV-1)-Infected Cells License Quiescent CD4 + T Lymphocytes to Replicate HIV-1 through a Nef- and ADAM17-Dependent Mechanism. J. Virol. 2014, 88, 11529–11539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avval, Z.M.; Malekpour, L.; Raeisi, F.; Babapoor, A.; Mousavi, S.M.; Hashemi, S.A.; Salari, M. Introduction of magnetic and supermagnetic nanoparticles in new approach of targeting drug delivery and cancer therapy application. Drug Metab. Rev. 2020, 52, 157–184. [Google Scholar] [CrossRef] [PubMed]
- Denzer, K.; Kleijmeer, M.; Heijnen, H.; Stoorvogel, W.; Geuze, H. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000, 113, 3365–3374. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Borja, M.; Wubbolts, R.; Calafat, J.; Janssen, H.; Divecha, N.; Dusseljee, S.; Neefjes, J. Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr. Biol. 1999, 9, 55–58. [Google Scholar] [CrossRef] [Green Version]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta (BBA) Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurunathan, S.; Kang, M.-H.; Kim, J.-H. A Comprehensive Review on Factors Influences Biogenesis, Functions, Therapeutic and Clinical Implications of Exosomes. Int. J. Nanomed. 2021, 16, 1281–1312. [Google Scholar] [CrossRef]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010, 117, 1–4. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Ramakrishna, S.; Esmaeili, H.; Bahrani, S.; Koosha, M.; Babapoor, A. Green synthesis of supermagnetic Fe3O4–MgO nanoparticles via Nutmeg essential oil toward superior anti-bacterial and anti-fungal performance. J. Drug Deliv. Sci. Technol. 2019, 54, 101352. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [Green Version]
- Bukong, T.N.; Momen-Heravi, F.; Kodys, K.; Bala, S.; Szabo, G. Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90. PLoS Pathog. 2014, 10, e1004424. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, S.; Fazilati, M.; Mousavi, S.M.; Nazem, H. Anti-bacterial/fungal and anti-cancer performance of green synthesized Ag nanoparticles using summer savory extract. J. Exp. Nanosci. 2020, 15, 363–380. [Google Scholar] [CrossRef]
- Dreux, M.; Garaigorta, U.; Boyd, B.; Décembre, E.; Chung, J.; Whitten-Bauer, C.; Wieland, S.; Chisari, F.V. Short-Range Exosomal Transfer of Viral RNA from Infected Cells to Plasmacytoid Dendritic Cells Triggers Innate Immunity. Cell Host Microbe 2012, 12, 558–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; He, Z.; Yuan, J.; Wen, W.; Huang, X.; Hu, Y.; Lin, C.; Pan, J.; Li, R.; Deng, H.; et al. IFITM3-containing exosome as a novel mediator for anti-viral response in dengue virus infection. Cell. Microbiol. 2015, 17, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Honegger, A.; Schilling, D.; Bastian, S.; Sponagel, J.; Kuryshev, V.; Sültmann, H.; Scheffner, M.; Hoppe-Seyler, K. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015, 11, e1004712. [Google Scholar] [CrossRef]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, S.; Fazilati, M.; Nazem, H.; Mousavi, S.M.; Ramoji, K. Green Synthesis of Magnetic Nanoparticles Using Satureja hortensis Essential Oil toward Superior Antibacterial/Fungal and Anticancer Performance. BioMed Res. Int. 2021, 2021, 1–14. [Google Scholar] [CrossRef]
- Friand, V.; David, G.; Zimmermann, P. Syntenin and syndecan in the biogenesis of exosomes. Biol. Cell 2015, 107, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Abootalebi, S.N.; Mousavi, S.M.; Hashemi, S.A.; Shorafa, E.; Omidifar, N.; Gholami, A. Antibacterial Effects of Green-Synthesized Silver Nanoparticles Using Ferula asafoetida against Acinetobacter baumannii Isolated from the Hospital Environment and Assessment of Their Cytotoxicity on the Human Cell Lines. J. Nanomater. 2021, 2021, 1–12. [Google Scholar] [CrossRef]
- Theos, A.C.; Truschel, S.T.; Tenza, D.; Hurbain, I.; Harper, D.C.; Berson, J.F.; Thomas, P.C.; Raposo, G.; Marks, M.S. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev. Cell 2006, 10, 343–354. [Google Scholar] [CrossRef] [Green Version]
- Mousavi, S.M.; Hashemi, S.A.; Gholami, A.; Omidifar, N.; Zarei, M.; Bahrani, S.; Babapoor, A. Bioinorganic synthesis of polyrhodanine stabilized Fe3O4/Graphene oxide in microbial supernatant media for anticancer and antibacterial applications. Bioinorg. Chem. Appl. 2021, 2021, 9972664. [Google Scholar] [CrossRef]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell Surface Tetraspanin Tspan8 Contributes to Molecular Pathways of Exosome-Induced Endothelial Cell Activation. Cancer Res. 2010, 70, 1668–1678, Erratum in Cancer Res. 2010, 70, 6683. [Google Scholar] [CrossRef] [Green Version]
- Masoumzadeh, R. Polyethyleneimine-based materials for gene therapy, bioimaging and drug delivery systems applications. Adv. Appl. NanoBio Technol. 2021, 2, 13–16. [Google Scholar]
- Vigna, E.; Naldini, L. Lentiviral vectors: Excellent tools for experimental gene transfer and promising candidates for gene therapy. J. Gene Med. 2000, 2, 308–316. [Google Scholar] [CrossRef]
- Manceur, A.P.; Kim, H.; Misic, V.; Andreev, N.; Dorion-Thibaudeau, J.; Lanthier, S.; Bernier, A.; Tremblay, S.; Gélinas, A.-M.; Broussau, S.; et al. Scalable Lentiviral Vector Production Using Stable HEK293SF Producer Cell Lines. Hum. Gene Ther. Methods 2017, 28, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Lévy, C.; Verhoeyen, E.; Cosset, F.-L. Surface engineering of lentiviral vectors for gene transfer into gene therapy target cells. Curr. Opin. Pharmacol. 2015, 24, 79–85. [Google Scholar] [CrossRef]
- Tokgün, O.; Fiorentino, F.P.; Tokgun, P.E.; Yokota, J.; Akca, H. Design of a Lentiviral Vector for the Inducible Expression of MYC: A New Strategy for Construction Approach. Mol. Biotechnol. 2017, 59, 200–206. [Google Scholar] [CrossRef]
- Kim, S.-H.; Samal, S.K. Newcastle Disease Virus as a Vaccine Vector for Development of Human and Veterinary Vaccines. Viruses 2016, 8, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhao, W.; Li, D.; Yang, J.; Zsak, L.; Yu, Q. Development of a Newcastle disease virus vector expressing a foreign gene through an internal ribosomal entry site provides direct proof for a sequential transcription mechanism. J. Gen. Virol. 2015, 96, 2028–2035. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.; Chen, C.; Guofu, C.; Zhang, Z.; Shang, Y.; Shao, H.; Luo, Q.; Yang, J.; Wang, H.; Wang, H.; et al. Development of a novel thermostable Newcastle disease virus vaccine vector for expression of a heterologous gene. J. Gen. Virol. 2015, 96, 1219–1228. [Google Scholar] [CrossRef]
- Fradet-Turcotte, A.; Bergeron-Labrecque, F.; Moody, C.A.; Lehoux, M.; Laimins, L.A.; Archambault, J. Nuclear Accumulation of the Papillomavirus E1 Helicase Blocks S-Phase Progression and Triggers an ATM-Dependent DNA Damage Response. J. Virol. 2011, 85, 8996–9012. [Google Scholar] [CrossRef] [Green Version]
- Kadaja, M.; Isok-Paas, H.; Laos, T.; Ustav, E.; Ustav, M. Mechanism of Genomic Instability in Cells Infected with the High-Risk Human Papillomaviruses. PLoS Pathog. 2009, 5, e1000397. [Google Scholar] [CrossRef]
- Reinson, T.; Toots, M.; Kadaja, M.; Pipitch, R.; Allik, M.; Ustav, E.; Ustav, M. Engagement of the ATR-Dependent DNA Damage Response at the Human Papillomavirus 18 Replication Centers during the Initial Amplification. J. Virol. 2013, 87, 951–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, S.; Luo, Y.; Cheng, F.; Huang, Q.; Shen, W.; Kleiboeker, S.; Qiu, J. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at phase G2/M. J. Virol. 2012, 86, 10748–10758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Kleiboeker, S.; Deng, X.; Qiu, J. Human Parvovirus B19 Infection Causes Cell Cycle Arrest of Human Erythroid Progenitors at Late S Phase That Favors Viral DNA Replication. J. Virol. 2013, 87, 12766–12775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Lou, S.; Deng, X.; Liu, Z.; Li, Y.; Kleiboeker, S.; Qiu, J. Parvovirus B19 Infection of Human Primary Erythroid Progenitor Cells Triggers ATR-Chk1 Signaling, Which Promotes B19 Virus Replication. J. Virol. 2011, 85, 8046–8055. [Google Scholar] [CrossRef] [Green Version]
- Usme-Ciro, J.A.; Campillo-Pedroza, N.; Almazán, F.; Gallego-Gomez, J.C. Cytoplasmic RNA viruses as potential vehicles for the delivery of therapeutic small RNAs. Virol. J. 2013, 10, 185. [Google Scholar] [CrossRef] [Green Version]
- Arabi, Y.M.; Hajeer, A.H.; Luke, T.; Raviprakash, K.; Balkhy, H.; Johani, S.; Al-Dawood, A.; Al-Qahtani, S.; Al-Omari, A.; Al-Hameedet, F.; et al. F.; et al. Feasibility of using convalescent plasma immunotherapy for MERS-CoV infection, Saudi Arabia. Emerg. Infect. Dis. 2016, 22, 1554. [Google Scholar] [CrossRef]
- Rojas, M.; Rodríguez, Y.; Monsalve, D.M.; Acosta-Ampudia, Y.; Camacho, B.; Gallo, J.E.; Rojas-Villarraga, A.; Ramírez-Santana, C.; Díaz-Coronado, J.C.; Manrique, R.; et al. Convalescent plasma in Covid-19: Possible mechanisms of action. Autoimmun. Rev. 2020, 19, 102554. [Google Scholar] [CrossRef]
- Takmil, F.; Esmaeili, H.; Mousavi, S.M.; Hashemi, S.A. Nano-magnetically modified activated carbon prepared by oak shell for treatment of wastewater containing fluoride ion. Adv. Powder Technol. 2020, 31, 3236–3245. [Google Scholar] [CrossRef]
- Marano, G.; Vaglio, S.; Pupella, S.; Facco, G.; Catalano, L.; Liumbruno, G.M.; Grazzini, G. Convalescent plasma: New evidence for an old therapeutic tool? Blood Transfus. 2016, 14, 152. [Google Scholar]
- Hung, I.F.N.; To, K.; Lee, C.-K.; Lee, K.-L.; Chan, K.K.C.; Yan, W.-W.; Liu, R.; Watt, C.-L.; Chan, W.-M.; Lai, K.-Y.; et al. Convalescent Plasma Treatment Reduced Mortality in Patients with Severe Pandemic Influenza A (H1N1) 2009 Virus Infection. Clin. Infect. Dis. 2011, 52, 447–456. [Google Scholar] [CrossRef]
- van Griensven, J.; De Weiggheleire, A.; Delamou, A.; Smith, P.G.; Edwards, T.; Vandekerckhove, P.; Lynen, L. The use of Ebola convalescent plasma to treat Ebola virus disease in resource-constrained settings: A perspective from the field. Clin. Infect. Dis. 2016, 62, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Hashemi, S.A.; Amani, A.M.; Saed, H.; Jahandideh, S.; Mojoudi, F. Polyethylene Terephthalate/Acryl Butadiene Styrene Copolymer Incorporated with Oak Shell, Potassium Sorbate and Egg Shell Nanoparticles for Food Packaging Applications: Control of Bacteria Growth, Physical and Mechanical Properties. Polym. Renew. Resour. 2017, 8, 177–196. [Google Scholar] [CrossRef]
- Yin, K.; Wang, S.; Zhao, R.C. Exosomes from mesenchymal stem/stromal cells: A new therapeutic paradigm. Biomark. Res. 2019, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pashoutan Sarvar, D.; Shamsasenjan, K.; Akbarzadehlaleh, P. Mesenchymal Stem Cell-Derived Exosomes: New Opportunity in Cell-Free Therapy. Adv. Pharm. Bull. 2016, 6, 293–299. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, S.A.; Mousavi, S.M.; Bahrani, S.; Ramakrishna, S. Integrated polyaniline with graphene oxide-iron tungsten nitride nanoflakes as ultrasensitive electrochemical sensor for precise detection of 4-nitrophenol within aquatic media. J. Electroanal. Chem. 2020, 873, 114406. [Google Scholar] [CrossRef]
- Zhou, Y.; Kosaka, N.; Xiao, Z.; Ochiya, T. MSC-Exosomes in Regenerative Medicine; Elsevier: Amsterdam, The Netherlands, 2020; pp. 433–465. [Google Scholar] [CrossRef]
- Pinky; Gupta, S.; Krishnakumar, V.; Sharma, Y.; Dinda, A.K.; Mohanty, S. Mesenchymal Stem Cell Derived Exosomes: A Nano Platform for Therapeutics and Drug Delivery in Combating COVID-19. Stem Cell Rev. Rep. 2021, 17, 33–43. [Google Scholar] [CrossRef]
- Azhdari, R.; Mousavi, S.M.; Hashemi, S.A.; Bahrani, S.; Ramakrishna, S. Decorated graphene with aluminum fumarate metal organic framework as a superior non-toxic agent for efficient removal of Congo Red dye from wastewater. J. Environ. Chem. Eng. 2019, 7, 103437. [Google Scholar] [CrossRef]
- Sengupta, V.; Sengupta, S.; Lazo, A.; Woods, P.; Nolan, A.; Bremer, N. Exosomes derived from bone marrow mesenchymal stem cells as treatment for severe COVID-19. Stem Cells Dev. 2020, 29, 747–754. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.-J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genom. 2020, 47, 119–121. [Google Scholar] [CrossRef]
- Xu, K.; Cai, H.; Shen, Y.; Ni, Q.; Chen, Y.; Hu, S.; Li, J.; Wang, H.; Yu, L.; Huang, H.; et al. Management of corona virus disease-19 (COVID-19): The Zhejiang experience. Zhejiang Da Xue Xue Bao. Yi Xue Ban 2020, 49, 147–157. [Google Scholar]
- Badierah, R.A.; Uversky, V.N.; Redwan, E.M. Dancing with Trojan horses: An interplay between the extracellular vesicles and viruses. J. Biomol. Struct. Dyn. 2021, 39, 3034–3060. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour, M.; Rezaie, J.; Nouri, M.; Panahi, Y. The role of extracellular vesicles in COVID-19 virus infection. Infect. Genet. Evol. 2020, 85, 104422. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Travanty, E.A.; Oko, L.; Edeen, K.; Berglund, A.; Wang, J.; Ito, Y.; Holmes, K.V.; Mason, R.J. Innate Immune Response of Human Alveolar Type II Cells Infected with Severe Acute Respiratory Syndrome–Coronavirus. Am. J. Respir. Cell Mol. Biol. 2013, 48, 742–748. [Google Scholar] [CrossRef] [Green Version]
- Knoops, K.; Kikkert, M.; Worm, S.H.E.V.D.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Kalashgrani, M.Y.; Omidifar, N.; Bahrani, S.; Rao, N.V.; Babapoor, A.; Gholami, A.; Chiang, W.-H. Bioactive Graphene Quantum Dots Based Polymer Composite for Biomedical Applications. Polymers 2022, 14, 617. [Google Scholar] [CrossRef]
- Kouwaki, T.; Okamoto, M.; Tsukamoto, H.; Fukushima, Y.; Oshiumi, H. Extracellular Vesicles Deliver Host and Virus RNA and Regulate Innate Immune Response. Int. J. Mol. Sci. 2017, 18, 666. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.-J.; Na, D.H.; Kwak, J.H.; Douaisi, M.; Zhang, F.; Park, E.J.; Park, J.-H.; Youn, H.; Song, C.-S.; Kane, R.S.; et al. Nanostructured glycan architecture is important in the inhibition of influenza A virus infection. Nat. Nanotechnol. 2017, 12, 48–54. [Google Scholar] [CrossRef]
- Alipour, A.; Kalashgarani, M.Y. Nano Protein and Peptides for Drug Delivery and Anticancer Agents. Adv. Appl. NanoBio-Technol. 2022, 3, 60–64. [Google Scholar]
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [Google Scholar] [CrossRef] [Green Version]
- Haselhorst, T.; Fleming, F.E.; Dyason, J.C.; Hartnell, R.D.; Yu, X.; Holloway, G.; Santegoets, K.C.M.; Kiefel, M.J.; Blanchard, H.; Coulson, B.; et al. Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 2009, 5, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Kalashgarani, M.Y.; Babapoor, A. Application of nano-antibiotics in the diagnosis and treatment of infectious diseases. Adv. Appl. NanoBio-Technol. 2022, 3, 22–35. [Google Scholar]
- Harden, M.E.; Munger, K. Human papillomavirus 16 E6 and E7 oncoprotein expression alters microRNA expression in extracellular vesicles. Virology 2017, 508, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Heo, D.S.; Snyderman, C.; Gollin, S.M.; Pan, S.; Walker, E.; Deka, R.; Barnes, E.L.; Johnson, J.T.; Herberman, R.B.; Whiteside, T.L. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer Res. 1989, 49, 5167–5175. [Google Scholar] [PubMed]
- Schuler, P.J.; Schilling, B.; Harasymczuk, M.; Hoffmann, T.K.; Johnson, J.; Lang, S.; Whiteside, T.L. Phenotypic and functional characteristics of CD4+ CD39+ FOXP3+ and CD4+ CD39+ FOXP3neg T-cell subsets in cancer patients. Eur. J. Immunol. 2012, 42, 1876–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, C.-S.; Funk, S.; Muller, L.; Boyiadzis, M.; Whiteside, T.L. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J. Extracell. Vesicles 2016, 5, 29289. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.-S.; Sharma, P.; Yerneni, S.S.; Simms, P.; Jackson, E.K.; Whiteside, T.L.; Boyiadzis, M. Circulating exosomes carrying an immunosuppressive cargo interfere with cellular immunotherapy in acute myeloid leukemia. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, L.; Mitsuhashi, M.; Simms, P.; Gooding, W.E.; Whiteside, T.L. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci. Rep. 2016, 6, 20254. [Google Scholar] [CrossRef] [Green Version]
- Wieckowski, E.U.; Visus, C.; Szajnik, M.; Szczepanski, M.J.; Storkus, W.J.; Whiteside, T.L. Tumor-Derived Microvesicles Promote Regulatory T Cell Expansion and Induce Apoptosis in Tumor-Reactive Activated CD8+T Lymphocytes. J. Immunol. 2009, 183, 3720–3730. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Campoli, M.; Cho, H.S.; Ogino, T.; Bandoh, N.; Shen, J.; Hur, S.Y.; Kageshita, T.; Ferrone, S. A method to generate antigen-specific mAb capable of staining formalin-fixed, paraffin-embedded tissue sections. J. Immunol. Methods 2005, 299, 139–151. [Google Scholar] [CrossRef]
- Theodoraki, M.-N.; Hoffmann, T.K.; Whiteside, T.L. Separation of plasma-derived exosomes into CD3(+) and CD3(−) fractions allows for association of immune cell and tumour cell markers with disease activity in HNSCC patients. Clin. Exp. Immunol. 2018, 192, 271–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welters, M.J.; Ma, W.; Santegoets, S.J.; Goedemans, R.; Ehsan, I.; Jordanova, E.S.; Van Ham, V.J.; Van Unen, V.; Koning, F.; Van Egmond, S.I.; et al. Intratumoral HPV16-Specific T Cells Constitute a Type I–Oriented Tumor Microenvironment to Improve Survival in HPV16-Driven Oropharyngeal Cancer. Clin. Cancer Res. 2018, 24, 634–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiteside, T.L. Head and Neck Carcinoma Immunotherapy: Facts and Hopes. Clin. Cancer Res. 2017, 24, 6–13. [Google Scholar] [CrossRef] [Green Version]
- Guenat, D.; Hermetet, F.; Prétet, J.-L.; Mougin, C. Exosomes and Other Extracellular Vesicles in HPV Transmission and Carcinogenesis. Viruses 2018, 9, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Liu, K.; Liu, Y.; Xu, Y.; Zhang, F.; Yang, H.; Liu, J.; Pan, T.; Chen, J.; Wu, M.; et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 2013, 14, 793–803. [Google Scholar] [CrossRef]
- Sun, C.; Fu, B.; Gao, Y.; Liao, X.; Sun, R.; Tian, Z.; Wei, H. TGF-β1 down-regulation of NKG2D/DAP10 and 2B4/SAP expression on human NK cells contributes to HBV persistence. PLoS Pathog. 2012, 8, e1002594. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, C.; Zhang, J. HMBOX1 negatively regulates NK cell functions by suppressing the NKG2D/DAP10 signaling pathway. Cell. Mol. Immunol. 2011, 8, 433–440. [Google Scholar] [CrossRef] [Green Version]
- Trippler, M.; Büschenfelde, K.-H.M.Z.; Gerken, G. HBV viral load within subpopulations of peripheral blood mononuclear cells in HBV infection using limiting dilution PCR. J. Virol. Methods 1999, 78, 129–147. [Google Scholar] [CrossRef]
- Lee, Z.; Nishikawa, S.; Gao, S.; Eksteen, J.B.; Czub, M.; Gill, M.J.; Osiowy, C.; van der Meer, F.; Van Marle, G.; Coffin, C.S. Detection of Hepatitis B Virus (HBV) Genomes and HBV Drug Resistant Variants by Deep Sequencing Analysis of HBV Genomes in Immune Cell Subsets of HBV Mono-Infected and/or Human Immunodeficiency Virus Type-1 (HIV-1) and HBV Co-Infected Individuals. PLoS ONE 2015, 10, e0137568. [Google Scholar] [CrossRef]
- Wang, J.; Horton, R.; Varthakavi, V.; Spearman, P.; Ratner, L. Formation and release of virus-like particles by HIV-1 matrix protein. AIDS 1999, 13, 281–283. [Google Scholar] [CrossRef]
- Cantin, R.; Diou, J.; Bélanger, D.; Tremblay, A.M.; Gilbert, C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods 2008, 338, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Usami, Y.; Popov, S.; Popova, E.; Inoue, M.; Weissenhorn, W.; Göttlinger, H.G. The ESCRT pathway and HIV-1 budding. Biochem. Soc. Trans. 2009, 37, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Chertova, E.; Chertov, O.; Coren, L.V.; Roser, J.D.; Trubey, C.M.; Bess, J.W.; Sowder, R.C.; Barsov, E.; Hood, B.L.; Fisher, R.J.; et al. Proteomic and Biochemical Analysis of Purified Human Immunodeficiency Virus Type 1 Produced from Infected Monocyte-Derived Macrophages. J. Virol. 2006, 80, 9039–9052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gould, S.; Booth, A.M.; Hildreth, J.E. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 2003, 100, 10592–10597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreibelt, G.; Bol, K.F.; Westdorp, H.; Wimmers, F.; Aarntzen, E.H.J.G.; Duiveman-de Boer, T.; Van De Rakt, M.W.M.M.; Scharenborg, N.M.; De Boer, A.J.; Pots, J.M.; et al. Effective Clinical Responses in Metastatic Melanoma Patients after Vaccination with Primary Myeloid Dendritic Cells. Clin. Cancer Res. 2016, 22, 2155–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Cosme, J.; Liu, P.P.; Gramolini, A.O. The cardiovascular exosome: Current perspectives and potential. Proteomics 2013, 13, 1654–1659. [Google Scholar] [CrossRef]
- He, Y.; Deng, F.; Yang, S.; Wang, D.; Chen, X.; Zhong, S.; Zhao, J.; Tang, J. Exosomal microRNA: A novel biomarker for breast cancer. Biomark. Med. 2018, 12, 177–188. [Google Scholar] [CrossRef]
- Haque, S.; Vaiselbuh, S. Silencing of Exosomal miR-181a Reverses Pediatric Acute Lymphocytic Leukemia Cell Proliferation. Pharmaceuticals 2020, 13, 241. [Google Scholar] [CrossRef]
- Chen, W.; Li, Z.; Wu, T.; Li, J.; Li, X.; Liu, L.; Bai, H.; Ding, S.; Li, X.; Yu, X. Surface plasmon resonance biosensor for exosome detection based on reformative tyramine signal amplification activated by molecular aptamer beacon. J. Nanobiotechnol. 2021, 19, 1–10. [Google Scholar] [CrossRef]
- Ren, J.; He, W.; Zheng, L.; Duan, H. From structures to functions: Insights into exosomes as promising drug delivery vehicles. Biomater. Sci. 2016, 4, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L.; Scott, M.J.; Wickline, S.A. Maximizing exosome colloidal stability following electroporation. Anal. Biochem. 2014, 448, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Duan, Y.; Adkins, G.B.; Pan, S.; Wang, H.; Liu, Y.; Zhong, W. Highly Efficient Exosome Isolation and Protein Analysis by an Integrated Nanomaterial-Based Platform. Anal. Chem. 2018, 90, 2787–2795. [Google Scholar] [CrossRef] [PubMed]
- Bosch, S.; De Beaurepaire, L.; Allard, M.; Mosser, M.; Heichette, C.; Chrétien, D.; Jegou, D.; Bach, J.-M. Trehalose prevents aggregation of exosomes and cryodamage. Sci. Rep. 2016, 6, 36162. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.K. Method for Lyophilising an Exosome. U.S. Patent 16/340,948, 1 August 2019. [Google Scholar]
- Wang, Q.; Yang, Q.; Wang, Z.; Tong, H.; Ma, L.; Zhang, Y.; Shan, F.; Meng, Y.; Yuan, Z. Comparative analysis of human mesenchymal stem cells from fetal-bone marrow, adipose tissue, and Warton’s jelly as sources of cell immunomodulatory therapy. Hum. Vaccines Immunother. 2016, 12, 85–96. [Google Scholar] [CrossRef]
- Munoz, J.L.; Bliss, S.A.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Delivery of functional anti-miR-9 by mesenchymal stem cell–derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol. Ther. Nucleic Acids 2013, 2, e126. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, G.; Zhang, Y.; Jiang, H.; Wang, W.; Zhao, D.; Hong, J.; Yu, H.; Qi, L. Mesenchymal stem cell-derived exosomal microRNA-133b suppresses glioma progression via Wnt/β-catenin signaling pathway by targeting EZH2. Stem Cell Res. Ther. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Bellavia, D.; Raimondo, S.; Calabrese, G.; Forte, S.; Cristaldi, M.; Patinella, A.; Memeo, L.; Manno, M.; Raccosta, S.; Diana, P.; et al. Interleukin 3-receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 2017, 7, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.-G. A Novel Nanoparticle Drug Delivery System: The Anti-inflammatory Activity of Curcumin Is Enhanced When Encapsulated in Exosomes. Mol. Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef] [PubMed]






| Smart Nanoexosome Source | Cargo and Loading Mechanism | Effect Observed | Ref. |
|---|---|---|---|
| Mesechymal Stem Cell | Anti-miR-9 (Transfection) | Reversal of chemoresistance | [168] |
| miR-133 b (Transfection) | Suppression of progression | [169] | |
| Paclitaxel (Incubation) | Growth inhibition of human pancreatic adenocarcinoma cell | [170] | |
| Dendritic Cell | BACE1 siRNA (Electroporation) | Knockdown of specific gene after specific siRNA delivery to the brain for AD | [171] |
| Doxorubicin (Electroporation) | Specific drug delivery to the tumor site and inhibited tumor growth | [172] | |
| HEK293T | BCR-ABL siRNA (Transfection) | Overcome pharmacological resistance in CML cells | [173] |
| Mouse lymphoma cell | Curcumin (Mixing) | Increase anti-inflammatory activity | [174] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mousavi, S.M.; Hashemi, S.A.; Gholami, A.; Kalashgrani, M.Y.; Vijayakameswara Rao, N.; Omidifar, N.; Hsiao, W.W.-W.; Lai, C.W.; Chiang, W.-H. Plasma-Enabled Smart Nanoexosome Platform as Emerging Immunopathogenesis for Clinical Viral Infection. Pharmaceutics 2022, 14, 1054. https://doi.org/10.3390/pharmaceutics14051054
Mousavi SM, Hashemi SA, Gholami A, Kalashgrani MY, Vijayakameswara Rao N, Omidifar N, Hsiao WW-W, Lai CW, Chiang W-H. Plasma-Enabled Smart Nanoexosome Platform as Emerging Immunopathogenesis for Clinical Viral Infection. Pharmaceutics. 2022; 14(5):1054. https://doi.org/10.3390/pharmaceutics14051054
Chicago/Turabian StyleMousavi, Seyyed Mojtaba, Seyyed Alireza Hashemi, Ahmad Gholami, Masoomeh Yari Kalashgrani, Neralla Vijayakameswara Rao, Navid Omidifar, Wesley Wei-Wen Hsiao, Chin Wei Lai, and Wei-Hung Chiang. 2022. "Plasma-Enabled Smart Nanoexosome Platform as Emerging Immunopathogenesis for Clinical Viral Infection" Pharmaceutics 14, no. 5: 1054. https://doi.org/10.3390/pharmaceutics14051054
APA StyleMousavi, S. M., Hashemi, S. A., Gholami, A., Kalashgrani, M. Y., Vijayakameswara Rao, N., Omidifar, N., Hsiao, W. W. -W., Lai, C. W., & Chiang, W. -H. (2022). Plasma-Enabled Smart Nanoexosome Platform as Emerging Immunopathogenesis for Clinical Viral Infection. Pharmaceutics, 14(5), 1054. https://doi.org/10.3390/pharmaceutics14051054