Hospital Effluents and Wastewater Treatment Plants: A Source of Oxytetracycline and Antimicrobial-Resistant Bacteria in Seafood
Abstract
:1. Introduction
1.1. OTC and AMR Bacteria
1.2. Techniques for Treatment of HWW
1.2.1. Chemical Process
1.2.2. Biological Process
1.2.3. Physical Process
1.2.4. Physiochemical Process
2. Materials and Methods
2.1. Search Strategy
- Articles’ publication years were between 2010 and 2021;
- The keywords AMR and OTC successfully combined with “hospital treated and untreated wastewater”, “seafood”, “human health”, “hospital effluents”, “wastewater treatment plant,” and “seafood contamination” in the title and abstract;
- The articles had to be scientific indexed papers only;
- Search was limited to research articles only.
- They were published in languages other than English;
- Articles that only an abstract were available;
- Articles that are not related to the studies were also excluded.
2.2. Data Extraction and Reporting
- Location, sample size (L), total samples, WWTP effluent, WWTP influent, and AMR genes detected;
- Types of tetracycline antibiotic and region of the study;
- Results, including a mean antibiotic concentration in hospital effluents and wastewater treatment plants.
3. Results and Discussion
3.1. Levels of OTC and AMR Bacteria in HWW
3.2. AMR Genes in Bacterial Genomes
3.3. Bacterial Resistance to Antimicrobials
3.4. Heavy Metals as AMR Genes’ Co-Regulators
3.5. Connecting OTC, Heavy Metals, and AMR Bacteria
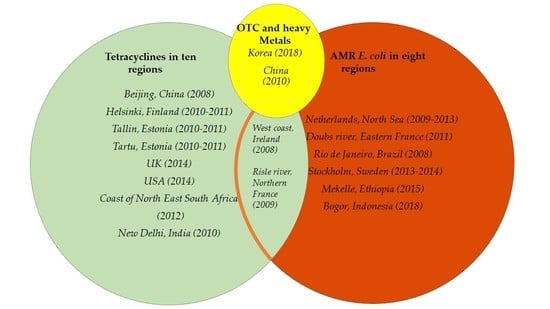
3.6. OTC and AMR Distribution Globally
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orias, F.; Perrodin, Y. Characterisation of the Ecotoxicity of Hospital Effluents: A Review. Sci. Total Environ. 2013, 454–455, 250–276. [Google Scholar] [CrossRef]
- Weissbrodt, D.; Kovalova, L.; Ort, C.; Pazhepurackel, V.; Moser, R.; Hollender, J.; Siegrist, H.; McArdell, C. Mass Flows of X-Ray Contrast Media and Cytostatics in Hospital Wastewater. Environ. Sci. Technol. 2009, 43, 4810–4817. [Google Scholar] [CrossRef] [Green Version]
- Emmanuel, E.; Keck, G.; Blanchard, J.; Vermande, P.; Perrodin, Y. Toxicological Effects of Disinfections Using Sodium Hypochlorite on Aquatic Organisms and Its Contribution to AOX Formation in Hospital Wastewater. Environ. Int. 2004, 30, 891–900. [Google Scholar] [CrossRef]
- Okoh, A.I.; Odjadjare, E.E.; Igbinosa, E.O.; Osode, A.N. Wastewater treatment plants as a source of microbial pathogens in receiving watersheds. Afr. J. Biotechnol. 2007, 6, 2932–2944. [Google Scholar] [CrossRef]
- Langford, K.H.; Thomas, K.V. Determination of Pharmaceutical Compounds in Hospital Effluents and Their Contribution to Wastewater Treatment Works. Environ. Int. 2009, 35, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Verlicchi, P.; Galletti, A.; Petrovic, M.; Barceló, D. Hospital Effluents as a Source of Emerging Pollutants: An Overview of Micropollutants and Sustainable Treatment Options. J. Hydrol. 2010, 389, 416–428. [Google Scholar] [CrossRef]
- Galvin, S.; Boyle, F.; Hickey, P.; Vellinga, A.; Morris, D.; Cormican, M. Enumeration and Characterization of Antimicrobial-Resistant Escherichia Coli Bacteria in Effluent from Municipal, Hospital, and Secondary Treatment Facility Sources. Appl. Environ. Microbiol. 2010, 76, 4772–4779. [Google Scholar] [CrossRef] [Green Version]
- Angulo, F.J.; Nargund, V.N.; Chiller, T.C. Evidence of an Association Between Use of Anti-Microbial Agents in Food Animals and Anti-Microbial Resistance Among Bacteria Isolated from Humans and the Human Health Consequences of Such Resistance. J. Vet. Med. Ser. B 2004, 51, 374–379. [Google Scholar] [CrossRef]
- Antimicrobial Resistance. Fact Sheet 194. World Health Organization. Available online: http://www.who.int/mediacentre/factsheets/fs194/en/ (accessed on 31 July 2021).
- The Medical Impact of the Use of Antimicrobials in Food Animals. Report of a WHO Meeting. Berlin, Germany, 13–17 October 1997. Available online: http://whqlibdoc.who.int/hq/1997/WHO_EMC_ZOO_97.4.pdf (accessed on 31 July 2021).
- de Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die A Year Due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [Green Version]
- Ekwanzala, M.D.; Dewar, J.B.; Kamika, I.; Momba, M.N. Tracking the Environmental Dissemination of Carbapenem-Resistant Klebsiella Pneumoniae Using Whole Genome Sequencing. Sci. Total Environ. 2019, 691, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Rose, W.E.; Rybak, M.J. Tigecycline: First of a New Class of Antimicrobial Agents. Pharmacotherapy 2006, 26, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Mihciokur, H.; Oguz, M. Removal of Oxytetracycline and Determining Its Biosorption Properties on Aerobic Granular Sludge. Environ. Toxicol. Pharmacol. 2016, 46, 174–182. [Google Scholar] [CrossRef]
- Ayandiran, T.O.; Falgenhauer, L.; Schmiedel, J.; Chakraborty, T.; Ayeni, F.A. High Resistance to Tetracycline and Ciprofloxacin in Bacteria Isolated from Poultry Farms in Ibadan, Nigeria. J. Infect. Dev. Ctries. 2018, 12, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infec. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogomolni, A.L.; Gast, R.J.; Ellis, J.C.; Dennett, M.; Pugliares, K.R.; Lentell, B.J.; Moore, M.J. Victims or Vectors: A Survey of Marine Vertebrate Zoonoses from Coastal Waters of the Northwest Atlantic. Dis. Aquat. Org. 2008, 81, 13–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grevskott, D.H.; Svanevik, C.S.; Sunde, M.; Wester, A.L.; Lunestad, B.T. Marine Bivalve Mollusks as Possible Indicators of Multidrug-Resistant Escherichia Coli and Other Species of the Enterobacteriaceae Family. Front. Microbiol. 2017, 8, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moller, T.S.; Overgaard, M.; Nielsen, S.S.; Bortolaia, V.; Sommer, M.O.; Guardabassi, L.; Olsen, J.E. Relation between Tetr and Teta Expression in Tetracycline Resistant Escherichia coli. BMC Microbiol. 2016, 16, 1–8. [Google Scholar] [CrossRef] [Green Version]
- D’Accolti, M.; Soffritti, I.; Mazzacane, S.; Caselli, E. Fighting AMR in the Healthcare Environment: Microbiome-Based Sanitation Approaches and Monitoring Tools. Int. J. Mol. Sci. 2019, 20, 1535. [Google Scholar] [CrossRef] [Green Version]
- Girijan, S.K.; Paul, R.; Rejish Kumar, V.J.; Pillai, D. Investigating the Impact of Hospital Antibiotic Usage on Aquatic Environment and Aquaculture Systems: A Molecular Study of Quinolone Resistance in Escherichia Coli. Sci. Total Environ. 2020, 748, 141538. [Google Scholar] [CrossRef] [PubMed]
- Jamborova, I.; Janecko, N.; Halova, D.; Sedmik, J.; Mezerova, K.; Papousek, I.; Kutilova, I.; Dolejska, M.; Cizek, A.; Literak, I. Molecular characterization of plasmid-mediated AmpC beta-lactamase- and extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae among corvids (Corvus brachyrhynchos and Corvus corax) roosting in Canada. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [Green Version]
- Lamba, M.; Graham, D.W.; Ahammad, S.Z. Hospital Wastewater Releases of Carbapenem-Resistance Pathogens and Genes in Urban India. Environ. Sci. Technol. 2017, 51, 13906–13912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaak, H.; Lynch, G.; Italiaander, R.; Hamidjaja, R.A.; Schets, F.M.; de Roda Husman, A.M. Multidrug-Resistant and Extended Spectrum Beta-Lactamase-Producing Escherichia Coli in Dutch Surface Water and Wastewater. PLoS ONE 2015, 10, e0127752. [Google Scholar] [CrossRef] [Green Version]
- Agwuh, K.N.; MacGowan, A. Pharmacokinetics and Pharmacodynamics of the Tetracyclines Including Glycylcyclines. J. Antimicrob. Chemother. 2006, 58, 256–265. [Google Scholar] [CrossRef] [Green Version]
- Rang, H.; Ritter, J.; Flower, R.; Henderson, G. Rang&Dale’s Pharmacology, 8th ed.; Elsevier Churchill Livingstone: London, UK, 2016; p. 619. [Google Scholar]
- Oberlé, K.; Capdeville, M.J.; Berthe, T.; Budzinski, H.; Petit, F. Evidence for a Complex Relationship Between Antibiotics and Antibiotic-Resistantescherichia Coli: From Medical Center Patients to a Receiving Environment. Environ. Sci. Technol. 2012, 46, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, J. Degradation of Norfloxacin in Aqueous Solution by Ionizing Irradiation: Kinetics, Pathway and Biological Toxicity. Chem. Eng. J. 2020, 395, 125095. [Google Scholar] [CrossRef]
- Wang, J.; Zhuan, R. Degradation of antibiotics by advanced oxidation processes: An overview. Sci. Total Environ. 2020, 701, 135023. [Google Scholar] [CrossRef] [PubMed]
- Paucar, N.; Kim, I.; Tanaka, H.; Sato, C. Ozone Treatment Process for the Removal of Pharmaceuticals and Personal Care Products in Wastewater. Ozone Sci. Eng. 2018, 41, 3–16. [Google Scholar] [CrossRef]
- Oh, J.; Salcedo, D.; Medriano, C.; Kim, S. Comparison of Different Disinfection Processes in the Effective Removal of Antibiotic-Resistant Bacteria and Genes. J. Environ. Sci. 2014, 26, 1238–1242. [Google Scholar] [CrossRef]
- Chen, J.; Deng, W.; Liu, Y.; Hu, L.; He, L.; Zhao, J.; Wang, T.; Ying, G. Fate and Removal of Antibiotics and Antibiotic Resistance Genes in Hybrid Constructed Wetlands. Environ. Pollut. 2019, 249, 894–903. [Google Scholar] [CrossRef]
- Furukawa, T.; Jikumaru, A.; Ueno, T.; Sei, K. Inactivation Effect of Antibiotic-Resistant Gene Using Chlorine Disinfection. Water 2017, 9, 547. [Google Scholar] [CrossRef]
- Jia, S.; Wu, J.; Ye, L.; Zhao, F.; Li, T.; Zhang, X. Metagenomic Assembly Provides A Deep Insight Into the Antibiotic Resistome Alteration Induced By Drinking Water Chlorination and Its Correlations With Bacterial Host Changes. J. Hazard. Mater. 2019, 379, 120841. [Google Scholar] [CrossRef]
- Destiani, R.; Templeton, M. Chlorination and Ultraviolet Disinfection of Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes in Drinking Water. AIMS Environ. Sci. 2019, 6, 222–241. [Google Scholar] [CrossRef]
- Majumder, A.; Gupta, A.; Ghosal, P.; Varma, M. A Review on Hospital Wastewater Treatment: A Special Emphasis on Occurrence and Removal of Pharmaceutically Active Compounds, Resistant Microorganisms, and SARS-Cov-2. J. Environ. Chem. Eng. 2021, 9, 104812. [Google Scholar] [CrossRef] [PubMed]
- Kosma, C.; Lambropoulou, D.; Albanis, T. Occurrence and Removal of Ppcps in Municipal and Hospital Wastewaters in Greece. J. Hazard. Mater. 2010, 179, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Jiang, X.; Xia, X.; Zhang, H.; Zheng, S. Detection, Occurrence and Fate of 22 Psychiatric Pharmaceuticals in Psychiatric Hospital and Municipal Wastewater Treatment Plants in Beijing, China. Chemosphere 2013, 90, 2520–2525. [Google Scholar] [CrossRef] [PubMed]
- Kappell, A.; Kimbell, L.; Seib, M.; Carey, D.; Choi, M.; Kalayil, T.; Fujimoto, M.; Zitomer, D.; McNamara, P. Removal of Antibiotic Resistance Genes in An Anaerobic Membrane Bioreactor Treating Primary Clarifier Effluent At 20 °C. Environ. Sci. Water Res. Technol. 2018, 4, 1783–1793. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Hong, P. Removal of Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes Affected By Varying Degrees of Fouling on Anaerobic Microfiltration Membranes. Environ. Sci. Technol. 2017, 51, 12200–12209. [Google Scholar] [CrossRef] [Green Version]
- Sui, Q.; Jiang, C.; Zhang, J.; Yu, D.; Chen, M.; Wang, Y.; Wei, Y. Does the Biological Treatment or Membrane Separation Reduce the Antibiotic Resistance Genes From Swine Wastewater Through A Sequencing-Batch Membrane Bioreactor Treatment Process. Environ. Int. 2018, 118, 274–281. [Google Scholar] [CrossRef]
- Bijlsma, L.; Pitarch, E.; Fonseca, E.; Ibanez, M.; Botero, A.; Claros, J.; Pastor, L.; Hernandez, F. Investigation of Pharmaceuticals in a Conventional Wastewater Treatment Plant: Removal Efficiency, Seasonal Variation and Impact of a Nearby Hospital. J. Environ. Chem. Eng. 2021, 9, 105548. [Google Scholar] [CrossRef]
- Yao, S.; Ye, J.; Yang, Q.; Hu, Y.; Zhang, T.; Jiang, L.; Munezero, S.; Lin, K.; Cui, C. Occurrence and Removal of Antibiotics, Antibiotic Resistance Genes, and Bacterial Communities in Hospital Wastewater. Environ. Sci. Pollut. Res. 2021, 28, 57321–57333. [Google Scholar] [CrossRef]
- Herraiz-Carboné, M.; Cotillas, S.; Lacasa, E.; Sainz de Baranda, C.; Riquelme, E.; Cañizares, P.; Rodrigo, M.; Sáez, C. A Review on Disinfection Technologies for Controlling the Antibiotic Resistance Spread. Sci. Total Environ. 2021, 797, 149150. [Google Scholar] [CrossRef]
- Grehs, B.; Lopes, A.; Moreira, N.; Fernandes, T.; Linton, M.; Silva, A.; Manaia, C.; Carissimi, E.; Nunes, O. Removal of Microorganisms and Antibiotic Resistance Genes From Treated Urban Wastewater: A Comparison Between Aluminium Sulphate and Tannin Coagulants. Water Res. 2019, 166, 115056. [Google Scholar] [CrossRef]
- Mathur, P.; Sanyal, D.; Callahan, D.; Conlan, X.; Pfeffer, F. Treatment Technologies to Mitigate the Harmful Effects of Recalcitrant Fluoroquinolone Antibiotics on the Environ- Ment and Human Health. Environ. Pollut. 2021, 291, 118233. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, F.; Sheng, H.; Xu, M.; Liang, Y.; Bian, Y.; Hashsham, S.; Jiang, X.; Tiedje, J. Enhanced Antibacterial Activity of Magnetic Biochar Conjugated Quaternary Phosphonium Salt. Carbon 2020, 163, 360–369. [Google Scholar] [CrossRef]
- Umar, M.; Roddick, F.; Fan, L. Moving From the Traditional Paradigm of Pathogen Inactivation to Controlling Antibiotic Resistance in Water—Role of Ultraviolet Irradiation. Sci. Total Environ. 2019, 662, 923–939. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiu, S.; Guo, J.; Ge, S. Light Irradiation Enables Rapid Start-Up of Nitritation through Suppressing Nxrb Gene Expression and Stimulating Ammonia-Oxidizing Bacteria. Environ. Sci. Technol. 2021, 55, 19. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.J.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Healthcare Interventions: Explanation and Elaboration. BMJ 2009, 339, b2700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Mutairi, K.A.; Yap, C.K. A Review of Heavy Metals in Coastal Surface Sediments from the Red Sea: Health-Ecological Risk Assessments. Int. J. Environ. Res. Public Health 2021, 18, 2798. [Google Scholar] [CrossRef]
- Armah, F.A.; Quansah, R.; Luginaah, I. A Systematic Review of Heavy Metals of Anthropogenic Origin in Environmental Media and Biota in the Context of Gold Mining in Ghana. Int. Sch. Res. Not. 2014, 2014, 1–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, A.; Xiao, Y.; Hu, J.; Asami, M.; Kunikane, S. Simultaneous Determination of Tetracyclines and Their Degradation Products in Environmental Waters By Liquid Chromatography–Electrospray Tandem Mass Spectrometry. J. Chromatogr. A 2009, 1216, 4655–4662. [Google Scholar] [CrossRef]
- Hou, J.; Wang, C.; Mao, D.; Luo, Y. The Occurrence and Fate of Tetracyclines in Two Pharmaceutical Wastewater Treatment Plants of Northern China. Environ. Sci. Pollut. Res. 2015, 23, 1722–1731. [Google Scholar] [CrossRef]
- Suzuki, S.; Ogo, M.; Koike, T.; Takada, H.; Newman, B. Sulfonamide and Tetracycline Resistance Genes in Total- and Culturable-Bacterial Assemblages in South African Aquatic Environments. Front. Microbiol. 2015, 6, 796. [Google Scholar] [CrossRef]
- Laht, M.; Karkman, A.; Voolaid, V.; Ritz, C.; Tenson, T.; Virta, M.; Kisand, V. Abundances of Tetracycline, Sulphonamide and Beta-Lactam Antibiotic Resistance Genes in Conventional Wastewater Treatment Plants (Wwtps) With Different Waste Load. PLoS ONE 2014, 9, e103705. [Google Scholar] [CrossRef] [Green Version]
- Borghi, A.A.; Palma, M.S. Tetracycline: Production, Waste Treatment and Environmental Impact Assessment. Braz. J. Pharm. Sci. 2014, 50, 25–40. [Google Scholar] [CrossRef]
- Chagas, T.P.; Seki, L.M.; Cury, J.C.; Oliveira, J.A.; Dávila, A.M.; Silva, D.M.; Asensi, M.D. Multiresistance, Beta-Lactamase-Encoding Genes and Bacterial Diversity in Hospital Wastewater in Rio De Janeiro, Brazil. J. Appl. Microbiol. 2011, 111, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Toltzis, P. 50 Years Ago in the Journal of Pediatrics. J. Pediatr. 2018, 197, 56. [Google Scholar] [CrossRef] [PubMed]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 26 May 2021).
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater Treatment Plants Release Large Amounts of Extended-Spectrum Β-Lactamase–Producing Escherichia Coli Into the Environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.K.; Colque, P.; Byfors, S.; Giske, C.G.; Möllby, R.; Kühn, I. Surveillance of Antimicrobial Resistance Among Escherichia Coli in Wastewater in Stockholm During 1 Year: Does It Reflect the Resistance Trends in the Society? Int. J. Antimicrob. Agents 2015, 4, 25–32. [Google Scholar] [CrossRef]
- Asfaw, T.; Negash, L.; Kahsay, A.; Weldu, Y. Antibiotic Resistant Bacteria From Treated and Untreated Hospital Wastewater At Ayder Referral Hospital, Mekelle, North Ethiopia. Adv. Microbiol. 2017, 7, 871–886. [Google Scholar] [CrossRef] [Green Version]
- Budiarti, S.; Lingga, R.; Rusmana, I.; Wahyudi, A.T. Antibiotics resistant Escherichia coli from hospital liquid waste. J. Appl. Biol. Sci. 2018, 1, 36–40. [Google Scholar]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 Positive Bacteria in the New Delhi Environment and Its Implications for Human Health: An Environmental Point Prevalence Study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus Epidermidis—The ‘Accidental’ Pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Herraiz-Carboné, M.; Cotillas, S.; Lacasa, E.; Sainz de Baranda, C.; Riquelme, E.; Cañizares, P.; Rodrigo, M.; Sáez, C. Are We Correctly Targeting the Research on Disinfection of Antibiotic-Resistant Bacteria (ARB)? J. Clean. Prod. 2021, 320, 128865. [Google Scholar] [CrossRef]
- Avcu, G.; Atıkan, B. Healthcare-Associated Infections At A Tertiary Level Pediatric Intensive Care Unit From Turkey. J. Pediatr. Res. 2021, 8, 246–250. [Google Scholar] [CrossRef]
- Ashagrie, D.; Genet, C.; Abera, B. Vancomycin-Resistant Enterococci and Coagulase-Negative Staphylococci Prevalence Among Patients Attending At Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Ethiopia. PLoS ONE 2021, 16, e0249823. [Google Scholar] [CrossRef]
- Hasanpour, F.; Neyestani, Z.; Arzanlou, M.; Moradi-Asl, E.; Sahebkar, A.; Khademi, F. Vancomycin-Resistant Enterococci in Iran: A Systematic Review and Meta-Analysis of Non-Clinical Studies. Gene Rep. 2021, 24, 101265. [Google Scholar] [CrossRef]
- Verlicchi, P. Trends, New Insights and Perspectives in the Treatment of Hospital Effluents. Curr. Opin. Environ. Sci. Health. 2021, 19, 100217. [Google Scholar] [CrossRef]
- Kuriyama, T.; Karasawa, T.; Williams, D. Antimicrobial Chemotherapy. Biofilms Infect. Prev. Control 2014, 209–244. [Google Scholar] [CrossRef]
- Browne, H.; Neville, B.; Forster, S.; Lawley, T. Transmission of the Gut Microbiota: Spreading of Health. Nat. Rev. Microbiol. 2017, 15, 531–543. [Google Scholar] [CrossRef] [Green Version]
- Verawaty, M.; Apriani, N.; Tarigan, L.R.; Apiran, E.T.; Laurenta, W.C. Muharni Antibiotics resistant Escherichia coli isolated from aquatic ecosystems in Palembang, South Sumatra, Indonesia. Biodiversitas 2020, 21, 46. [Google Scholar] [CrossRef]
- Iweriebor, B.C.; Gaqavu, S.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Antibiotic Susceptibilities of Enterococcus Species Isolated from Hospital and Domestic Wastewater Effluents in Alice, Eastern Cape Province of South Africa. Int. J. Environ. Res. Public Health 2015, 12, 4231–4246. [Google Scholar] [CrossRef]
- Omar Faruk, S.M.; Masudul Azad, C.A.M.; Nayeem, U.K. Isolation of Cefixime Resistant Salmonella from Hospitals Waste and Profiling Multi-Drug Resistance Pattern of the Selected Isolates. Int. Res. J. Biol. Sci. 2014, 3, 86–92. [Google Scholar]
- Chen, X.; Yin, H.; Li, G.; Wang, W.; Wong, P.; Zhao, H.; An, T. Antibiotic-Resistance Gene Transfer in Antibiotic-Resistance Bacteria Under Different Light Irradiation: Implications From Oxidative Stress and Gene Expression. Water Res. 2019, 149, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.; Schreier, H.; Lanska, L.; Hale, M. the Rising Tide of Antimicrobial Resistance in Aquaculture: Sources, Sinks and Solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, C.; Gin, K.Y. Monitoring Antimicrobial Resistance Dissemination in Aquatic Systems. Water 2019, 11, 71. [Google Scholar] [CrossRef] [Green Version]
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102. [Google Scholar] [CrossRef]
- Reinthaler, F.F.; Posch, J.; Feierl, G.; Wust, G.; Haas, D.; Ruckenbauer, G.; Mascher, F.; Marth, E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003, 37, 1685–1690. [Google Scholar] [CrossRef]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Bürgmann, H. Increased Levels of Multiresistant Bacteria and Resistance Genes after Wastewater Treatment and Their Dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Q.B.; Guo, M.T.; Yang, J. Fate of Antibiotic Resistant Bacteria and Genes during Wastewater Chlorination: Implication for Antibiotic Resistance Control. PLoS ONE 2015, 1, e0119403. [Google Scholar] [CrossRef]
- Liu, J.; Du, J.; Yi, L. Ra Tracer-Based Study of Submarine Groundwater Discharge and Associated Nutrient Fluxes into the Bohai Sea, China: A Highly Human-Affected Marginal Sea. J. Geophys. Res. Oceans 2017, 122, 8646–8660. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Madu, C.E.; Aiyegoro, O.A.; Stenström, T.A.; Okoh, A.I. Antibiogram and beta-lactamase genes among cefotaxime resistant E. coli from wastewater treatment plant. Antimicrob. Resist. Infect. Control 2020, 9, 46. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, A.Z.; Cen, T.; Li, X.; He, M.; Li, D.; Chen, J. Sub-Inhibitory Concentrations of Heavy Metals Facilitate the Horizontal Transfer of Plasmid-Mediated Antibiotic Resistance Genes in Water Environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Y.; Jin, M.; Yuan, Z.; Bond, P.; Guo, J. Both Silver Ions and Silver Nanoparticles Facilitate the Horizontal Transfer of Plasmid-Mediated Antibiotic Resistance Genes. Water Res. 2020, 169, 115229. [Google Scholar] [CrossRef]
- Mackulak, T.; Cverenkarova, K.; Vojs Stanova, A.; Feher, M.; Tamas, M.; Skulcova, A.; Gal, M.; Naumowicz, M.; Spalkova, V.; Birosova, L. Hospital Wastewater—Source of Specific Micropollutants, Antibiotic-Resistant Microorganisms, Viruses, and Their Elimination. Antibiotics 2021, 10, 1070. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, X.; Lang, X.; Qiao, X.; Li, X.; Chen, J. Insights into aquatic toxicities of the antibiotics oxytetracycline and ciprofloxacin in the presence of metal: Complexation versus mixture. Environ. Pollut. 2012, 166, 48–56. [Google Scholar] [CrossRef]
- Gao, M.; Lv, M.; Han, M.; Song, W.; Wang, D. Avoidance behavior of Eisenia fetida in oxytetracycline- and heavy metal-contaminated soils. Environ. Toxicol. Pharmacol. 2016, 47, 119–123. [Google Scholar] [CrossRef]
- Dahanayake, P.S.; Hossain, S.; Wickramanayake, M.V.; Heo, G.J. Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam (Ruditapes philippinarum) in Korea. J. Appl. Microbiol. 2019, 127, 941–952. [Google Scholar] [CrossRef]
- Zhang, H.; He, H.; Chen, S.; Huang, T.; Lu, K.; Zhang, Z.; Wang, R.; Zhang, X.; Li, H. Abundance of antibiotic resistance genes and their association with bacterial communities in activated sludge of wastewater treatment plants: Geographical distribution and network analysis. J. Environ. Sci. 2019, 82, 24–38. [Google Scholar] [CrossRef]
- Obayashi, Y.; Kadoya, A.; Kataoka, N.; Kanda, K.; Bak, S.M.; Iwata, H.; Suzuki, S. Tetracycline Resistance Gene Profiles in Red Seabream (Pagrus major) Intestine and Rearing Water After Oxytetracycline Administration. Front. Microbiol. 2020, 11, 1764. [Google Scholar] [CrossRef]
- Meehl, G.A. Characteristics of Surface Current Flow Inferred from a Global Ocean Current Data Set. J. Phys. Oceanogr. 1982, 12, 538–555. [Google Scholar] [CrossRef] [Green Version]
- GOV. UK National Statistics Sea Fisheries Annual Statistics Report 2019. Available online: https://www.gov.uk/government/statistics/uk-sea-fisheries-annual-statistics-report-2019 (accessed on 26 May 2021).
- UK Parliament House of Commons Library. Available online: https://commonslibrary.parliament.uk/research-briefings/sn02788/ (accessed on 26 May 2021).
- Spanggaard, B.; Jørgensen, F.; Gram, L.; Huss, H.H. Antibiotic resistance in bacteria isolated from three freshwater fish farms and an unpolluted stream in Denmark. Aquaculture 1993, 115, 195–207. [Google Scholar] [CrossRef]
- Henry, F.; Amara, R.; Courcot, L.; Lacouture, D.; Bertho, M.L. Heavy metals in four fish species from the French coast of the Eastern English Channel and Southern Bight of the North Sea. Environ. Int. 2004, 30, 675–683. [Google Scholar] [CrossRef]
- Reverter, M.; Sarter, S.; Caruso, D.; Avarre, J.C.; Combe, M.; Pepey, E.; Pouyaud, L.; Vega-Heredía, S.; de Verdal, H.; Gozlan, R.E. Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 2020, 11, 1870. [Google Scholar] [CrossRef] [Green Version]


| Antibiotic | Mean Antibiotic Concentration (ng/L) | AMR Bacteria or ARG | Matrix | Region | Reference |
|---|---|---|---|---|---|
| TC | N/D | Resistant E. coil 3264 cfu/100 mL | Hospital effluent | West coast, Ireland | [7] |
| TC | 10 | N/D | Hospital effluent | Risle river, Northern France | [27] |
| TC OTC | 1.9 3.8 | N/D | WWTP municipal Effluent | Beijing, China | [53] |
| OTC TC | 32.0 × 107 OTC 2.6 × 106 TC | N/D | PWWTP Influent and effluent | North China | [54] |
| TIG | N/D | blaNDM1 | Seepage and tap water | New Deli, India | [35] |
| OTC | N/D | 4.8 × 105 cfu/100 mL in river | River, WWTP, and surface water | Coast of North- East South Africa | [55] |
| 4.8 × 106 cfu/100 mL in WWTP | |||||
| tetM detected in surface water | |||||
| TC | N/D | tetM detected in 100% bacteria in all three locations tetC detected in 80% bacteria Helsinki, 27% Tallin, and 73% Tartu | WWTP effluent | Helsinki, Finland Tallin, Estonia Tartu, Estonia | [56] |
| OTC | 70–1340 ng/L | N/D | Surface water | USA | [57] |
| OTC | up to 340 ng/L | N/D | Surface water | UK | [57] |
| OTC | 71,700 ng/L | N/D | Runoff | England | [57] |
| Location | Sample Size (L) | Total Samples | WWTP Effluent | WWTP Influent | AMR Genes Detected | Reference |
|---|---|---|---|---|---|---|
| West coast, Ireland | 1 | 44 | 17 | 0 | blaCTX-M (blaCTX-M-28, blaCTX-M-3, blaCTX-M-61, blaCTX-M-15 blaCTX-M-14), blaTEM, blaSHV | [7] |
| Netherlands (North Sea) | 1 | 5 | 5 | 0 | blaOXA | [24] |
| Doubs river, Besancon, Eastern France | N/D | 1 | C | 1 | blaSHV | [61] |
| Risle river, Northern France | 1 | 48 1 | 24 1 | 24 1 | blaTEM | [27] |
| Rio de Janeiro, Brazil | 1 | 3 | 0 | 8 | blaCTX-M | [58] |
| Stockholm, Sweden | 0.05 | 6 | 6 | 0 | (blaCTX−M group1, blaCTX−M group9, blaCTX−M group2) | [62] |
| Mekelle, Ethiopia 2 | 0.125 & 0.25 | 20 | 20 | 0 | blaSHV, | [63] |
| Bogor, Indonesia | 0.25 | 1 | 0 | 1 | blaTEM | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCarthy, B.; Apori, S.O.; Giltrap, M.; Bhat, A.; Curtin, J.; Tian, F. Hospital Effluents and Wastewater Treatment Plants: A Source of Oxytetracycline and Antimicrobial-Resistant Bacteria in Seafood. Sustainability 2021, 13, 13967. https://doi.org/10.3390/su132413967
McCarthy B, Apori SO, Giltrap M, Bhat A, Curtin J, Tian F. Hospital Effluents and Wastewater Treatment Plants: A Source of Oxytetracycline and Antimicrobial-Resistant Bacteria in Seafood. Sustainability. 2021; 13(24):13967. https://doi.org/10.3390/su132413967
Chicago/Turabian StyleMcCarthy, Bozena, Samuel Obeng Apori, Michelle Giltrap, Abhijnan Bhat, James Curtin, and Furong Tian. 2021. "Hospital Effluents and Wastewater Treatment Plants: A Source of Oxytetracycline and Antimicrobial-Resistant Bacteria in Seafood" Sustainability 13, no. 24: 13967. https://doi.org/10.3390/su132413967
APA StyleMcCarthy, B., Apori, S. O., Giltrap, M., Bhat, A., Curtin, J., & Tian, F. (2021). Hospital Effluents and Wastewater Treatment Plants: A Source of Oxytetracycline and Antimicrobial-Resistant Bacteria in Seafood. Sustainability, 13(24), 13967. https://doi.org/10.3390/su132413967