An Approach for Foliar Trait Retrieval from Airborne Imaging Spectroscopy of Tropical Forests
Abstract
:1. Introduction
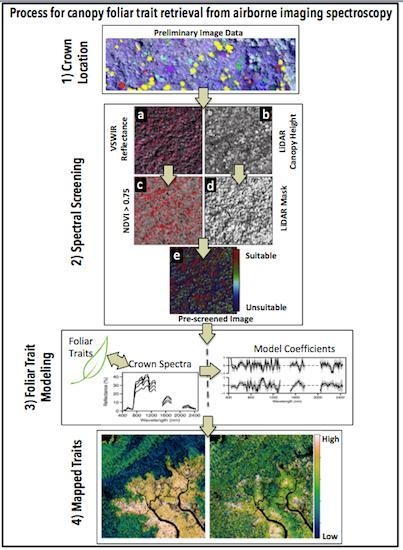
2. Materials and Methods
2.1. Study Region
2.2. Imaging Spectrometer Data
2.3. Foliar Sampling
2.4. Laboratory Assays
2.5. Linking Field and Remotely Sensed Data
3. Results
3.1. Canopy Chemical Variation
3.2. Airborne Spectral Variation
3.3. Model Calibration and Testing
4. Discussion
4.1. Spectral Quality
4.2. Model Development and Testing
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Townsend, A.R.; Asner, G.P.; Cleveland, C.C. The biogeochemical heterogeneity of tropical forests. Trends Ecol. Environ. 2008, 23, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Reichstein, M.; Bahn, M.; Mahecha, M.D.; Kattge, J.; Baldocchi, D.D. Linking plant and ecosystem functional biogeography. Proc. Natl. Acad. Sci. USA 2014. [Google Scholar] [CrossRef] [PubMed]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Ollinger, S.V. Sources of variability in canopy reflectance and the convergent properties of plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Kryston, K.; Vaughn, N.; Knapp, D.E.; Bentley, L.P.; Shenkin, A.; Salinas, N.; Sinca, F.; et al. Scale dependence of canopy trait distributions along a tropical forest elevation gradient. New Phytol. 2017, 214, 973–988. [Google Scholar] [CrossRef] [PubMed]
- Ustin, S.L. Remote sensing of canopy chemistry. Proc. Natl. Acad. Sci. USA 2013, 110, 804–805. [Google Scholar] [CrossRef] [PubMed]
- Homolova, L.; Malenovský, Z.; Clevers, J.G.; García-Santos, G.; Schaepman, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef]
- Kattge, J.; Diaz, S.; Lavorel, S.; Prentice, I.C.; Leadley, P.; Bonisch, G.; Garnier, E.; Westoby, M.; Reich, P.B.; Wright, I.J.; et al. TRY—A global database of plant traits. Glob. Chang. Biol. 2011, 17, 2905–2935. [Google Scholar] [CrossRef] [Green Version]
- Schimel, D.S.; Asner, G.P.; Moorcroft, P.R. Observing changing ecological diversity in the Anthropocene. Front. Ecol. Environ. 2013, 11, 129–137. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectranomics: Emerging science and conservation opportunities at the interface of biodiversity and remote sensing. Glob. Ecol. Conserv. 2016, 8, 212–219. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Anderson, C.B.; Martin, R.E.; Vaughn, N. Large-scale climatic and geophysical controls on the leaf economics spectrum. Proc. Natl. Acad. Sci. USA 2016, 113, 4043–4051. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Carranza-Jiménez, L.; Sinca, F.; Tupayachi, R.; Anderson, C.B.; Martinez, P. Functional and biological diversity of foliar spectra in tree canopies throughout the Andes to Amazon region. New Phytol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Knapp, D.E.; Boardman, J.; Green, R.O.; Kennedy-Bowdoin, T.; Eastwood, M.; Martin, R.E.; Anderson, C.; Field, C.B. Carnegie Airborne Observatory-2: Increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sens. Environ. 2012, 124, 454–465. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying forest canopy traits: Imaging spectroscopy versus field survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Vaughn, N.R.; Llactayo, W. Airborne laser-guided imaging spectroscopy to map forest trait diversity and guide conservation. Science 2017, 355, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, K.; Asner, G. Organismic-Scale Remote Sensing of Canopy Foliar Traits in Lowland Tropical Forests. Remote Sens. 2016, 8, 87. [Google Scholar] [CrossRef]
- Ashton, P.S. Dipterocarp biology as a window to the understanding of tropical forest structure. Ann. Rev. Ecol. Syst. 1988, 19, 347–370. [Google Scholar] [CrossRef]
- English, M.; Gillespie, G.; Ancrenaz, M.; Ismail, S.; Goossens, B.; Nathan, S.; Linklater, W. Plant selection and avoidance by the Bornean elephant (Elephas maximus borneensis) in tropical forest: Does plant recovery rate after herbivory influence food choices? J. Trop. Ecol. 2014, 30, 371–379. [Google Scholar] [CrossRef]
- Marsh, C.W.; Greer, A.G. Forest land-use in Sabah, Malaysia: An introduction to Danum Valley. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1992, 335, 331–339. [Google Scholar] [CrossRef]
- Born, J.; Pluess, A.R.; Burslem, D.F.; Nilus, R.; Maycock, C.R.; Ghazoul, J. Differing Life History Characteristics Support Coexistence of Tree Soil Generalist and Specialist Species in Tropical Rain Forests. Biotropica 2014, 46, 58–68. [Google Scholar] [CrossRef]
- Walsh, R.P.D.; Newbery, D.M. The ecoclimatology of Danum, Sabah, in the context of the world’s rainforest regions, with particular reference to dry periods and their impact. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Kitayama, K. An altitudinal transect study of the vegetation on Mount Kinabalu, Borneo. Vegetatio 1992, 102, 149–171. [Google Scholar] [CrossRef]
- Aiba, S.; Kitayama, K. Structure, composition and species diversity in an altitude-substrate matrix of rain forest tree communities on Mount Kinabalu, Borneo. Plant Ecol. 1999, 140, 139–157. [Google Scholar] [CrossRef]
- Kitayama, K.; Majalap-Lee, N.; Aiba, S. Soil phosphorus fractionation and phosphorus-use efficiencies of tropical rainforests along altitudinal gradients of Mount Kinabalu, Borneo. Oecologia 2000, 123, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.; Martin, R. Spectral and chemical analysis of tropical forests: Scaling from leaf to canopy levels. Remote Sens. Environ. 2008, 112, 3958–3970. [Google Scholar] [CrossRef]
- Colgan, M.S.; Baldeck, C.A.; Féret, J.B.; Asner, G.P. Mapping savanna tree species at ecosystem scales using support vector machine classification and BRDF correction on airborne hyperspectral and LiDAR data. Remote Sens. 2012, 4, 3462–3480. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Gillespie, K.M. Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin Coicalteau reagent. Nat. Protoc. 2007, 2, 875–877. [Google Scholar] [CrossRef] [PubMed]
- Toth, G.B.; Pavia, H. Removal of dissolved brown algal phlorotannins using insoluble Polyvinylpyrrolidone (PVPP). J. Chem. Ecol. 2001, 27, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- Feilhauer, H.; Asner, G.P.; Martin, R.E.; Schmidtlein, S. Brightness-normalized Partial Least Squares Regression for hyperspectral data. J. Quant. Spectrosc. Radiat. Transf. 2010, 111, 1947–1957. [Google Scholar] [CrossRef]
- Haaland, D.M.; Thomas, E.V. Partial least-squares methods for spectral Analyses. 1. Relation to other quantitative calibration methods and the extraction of qualitative information. Anal. Chem. 1988, 60, 1193–1202. [Google Scholar] [CrossRef]
- Martens, H. Reliable and relevant modelling of real world data: A personal account of the development of PLS Regression. Chemometr. Intell. Lab. Syst. 2001, 58, 85–95. [Google Scholar] [CrossRef]
- Chen, S.; Hong, X.; Harris, C.J.; Sharkey, P.M. Sparse modeling using orthogonal forest regression with PRESS statistic and regularization. IEEE Trans. Syst. Man Cybern. 2004, 34, 898–911. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Development Core Team, R. R: A Language and Environment For Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Schmidtlein, S.; Feilhauer, H.; Bruelheide, H. Mapping plant strategy types using remote sensing. J. Veg. Sci. 2012, 23, 395–405. [Google Scholar] [CrossRef]
- Martin, M.E.; Plourde, L.C.; Ollinger, S.V.; Smith, M.-L.; McNeil, B.E. A generalizable method for remote sensing of canopy nitrogen across a wide range of forest ecosystems. Remote Sens. Environ. 2008, 112, 3511–3519. [Google Scholar] [CrossRef]
- Roberts, D.A.; Ustin, S.L.; Ogunjemiyo, S.; Greenberg, J.; Dobrowski, S.Z.; Chen, J.Q.; Hinckley, T.M. Spectral and structural measures of northwest forest vegetation at leaf to landscape scales. Ecosystems 2004, 7, 545–562. [Google Scholar] [CrossRef]
- Mercier, G.; Lennon, M. Support vector machines for hyperspectral image classification with spectral-based kernels. In Proceedings of the 2003 IEEE International Geoscience and Remote Sensing Symposium, Toulouse, France, 21–25 July 2003; Volume 1, pp. 288–290. [Google Scholar]
- Knox, N.M.; Skidmore, A.K.; Sclerf, M.; de Boer, W.F.; van der Wall, C.; Prins, H.H.T.; Slotow, R. Nitrogen predition in grasses: Effect of bandwidth and plant material state on absorption feature selection. Int. J. Remote Sens. 2010, 31, 691–704. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E. Convergent elevation trends in canopy chemical traits of tropical forests. Glob. Chang. Biol. 2016, 22, 2216–2227. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Suhaili, A. Bin Sources of canopy chemical and spectral diversity in lowland Bornean forest. Ecosystems 2012, 15, 504–517. [Google Scholar] [CrossRef]
- Sterner, R.W.; Elser, J.J. Ecological Stoichiometry; Princeton University Press: Princeton, NJ, USA, 2002. [Google Scholar]
- Diaz, S.; Kattge, J.; Cornelissen, J.H.; Wright, I.J.; Lavorel, S.; Dray, S.; Reu, B.; Kleyer, M.; Wirth, C.; Prentice, I.C.; et al. The global spectrum of plant form and function. Nature 2016, 529, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Katabuchi, M.; Kurokawa, H.; Davies, S.J.; Tan, S.; Nakashizuka, T. Soil resource availability shapes community trait structure in a species-rich dipterocarp forest. J. Ecol. 2012, 100, 643–651. [Google Scholar] [CrossRef]
- Blackburn, G.A. Remote sensing of forest pigments using airborne imaging spectrometer and LIDAR imagery. Remote Sens. Environ. 2002, 82, 311–321. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Asner, G.; Martin, R. Spectroscopic Remote Sensing of Non-Structural Carbohydrates in Forest Canopies. Remote Sens. 2015, 7, 3526–3547. [Google Scholar] [CrossRef]
- Porter, L.J. Structure and Chemical Properties of the Condensed Tannins. In Plant Polyphenols; Springer: Boston, MA, USA, 1992; pp. 245–258. [Google Scholar]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Potts, M.D.; Ashton, P.S.; Kaufman, L.S.; Plotkin, J.B. Habitat Patterns in Tropical Rain Forests: A Comparison of 105 Plots in Northwest Borneo. Ecology 2012, 83, 2782–2797. [Google Scholar] [CrossRef]
- McGroddy, M.E.; Daufresne, T.; Hedin, L.O. Scaling of C:N:P stoichiometry in forests worldwide: Implications of terrestrial Redfield-type ratios. Ecology 2004, 85, 2390–2401. [Google Scholar] [CrossRef]
- Townsend, A.R.; Cleveland, C.C.; Asner, G.P.; Bustamante, M.M.C. Controls over foliar N:P ratios in tropical rain forests. Ecology 2007, 88, 107–118. [Google Scholar] [CrossRef]
- Asner, G.P. CHAPTER TWELVE A chemical-evolutionary basis for remote sensing of tropical forest diversity. In Forests and Global Change; Coomes, D.A., Barslem, D.F.R.P., Simonson, W.D., Eds.; Cambridge University Press: Cambridge, UK, 2014; pp. 343–358. [Google Scholar]
- Townsend, P.A.; Serbin, S.P.; Kruger, E.L.; Gamon, J.A. Disentangling the contribution of biological and physical properties of leaves and canopies in imaging spectroscopy data. Proc. Natl. Acad. Sci. USA 2013, 110, 4431–4432. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.G.P. Biophysical and biochemical sources of variability in canopy reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Asner, G.P.; Warner, A.S. Canopy shadow in IKONOS satellite observations of tropical forests and savannas. Remote Sens. Environ. 2003, 87, 521–533. [Google Scholar] [CrossRef]






| Site | Elev (m) | Substrate | Soil Order * | Crowns | Spectra | MAP mm·year−1 | MAT °C | N:P |
|---|---|---|---|---|---|---|---|---|
| Kinabatangan | 17 | Alluvial | Ult | 44 | 426 | 2883 | 27 | 15.3 |
| Danum Valley | 205 | Sedimentary | Ult | 76 | 1221 | 2882 | 26.7 | 19.7 |
| Sepilok | ||||||||
| 30 | Mudstone | Spod | 35 | 418 | 2975 | 27.2 | 18.7 | |
| 73 | Sandstone | Spod/Lit | 14 | 193 | 2975 | 27.2 | 27.3 | |
| 135 | Heath | Spod | 13 | 172 | 2975 | 27.2 | 34.0 | |
| Mt. Kinabalu | ||||||||
| 700 | Sedimentary | Oxi/Ult | 58 | 530 | 2509 | 23.9 | 19.0 | |
| 1700 | Sedimentary | Inc/Spod | 49 | 236 | 2718 | 18.9 | 24.3 | |
| 2700 | Sedimentary | Inc/Spod | 22 | 77 | 2085 | 13.3 | 29.1 | |
| 3100 | Granite | Inc. | 9 | 10 | 3285 | 10.6 | 21.8 | |
| 700 | Ultramafic | Oxi/Ult | 51 | 613 | 2509 | 23.7 | 26.8 | |
| 1700 | Ultramafic | Inc/Spod | 18 | 40 | 2718 | 17.3 | 31.2 | |
| 2700 | Ultramafic | Inc/Spod | 17 | 32 | 2085 | 12.7 | 25.8 | |
| 3100 | Ultramafic | Inc | 18 | 25 | 3285 | 10.7 | 31.4 |
| Model Development | Model Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Calibration | Validation | |||||||
| Trait | R2 | RMSE (%) | Latent Vectors | R2 | RMSE (%) | Trait Range | R2 | RMSE (%) |
| Light Capture and Growth | ||||||||
| LMA (g·m−2) | 0.79 | 27.3 (21.2) | 15 | 0.71 | 31.8 (24.7) | 127.5; 28.6–296.4 | 0.81 | 23.9 (21.6) |
| N (%) | 0.56 | 0.40 (22.7) | 20 | 0.46 | 0.45 (25.2) | 1.79; 0.60–4.46 | 0.54 | 0.43 (24.4) |
| Water (%) | 0.50 | 5.0 (8.8) | 16 | 0.32 | 5.8 (10.2) | 56.6; 38.2–79.9 | 0.40 | 5.2 (9.3) |
| Chl ab (mg·g−1) * | 0.43 | 1.32 (23.1) | 21 | 0.17 | 1.57 (27.6) | 5.6; 2.0–10.3 | 0.21 | 1.68 (29.8) |
| NSC (%) | 0.42 | 7.3 (16.9) | 14 | 0.16 | 8.7 (20.2) | 43.1; 23.1–75.1 | 0.22 | 7.8 (17.9) |
| δ13C (‰) | 0.38 | 1.2 (4.1) | 17 | 0.18 | 1.4 (4.8) | −30.0; −33.9–−24.5 | 0.38 | 1.2 (4.1) |
| Rock-Derived Nutrients | ||||||||
| P (%) | 0.55 | 0.03 (39.2) | 19 | 0.44 | 0.04 (43.4) | 0.09; 0.02–0.32 | 0.65 | 0.03 (36.6) |
| Ca (%) | 0.54 | 0.31 (50.4) | 16 | 0.39 | 0.36 (57.6) | 0.61; 0.06–2.72 | 0.49 | 0.29 (58.0) |
| K (%) | 0.42 | 0.29 (43.3) | 19 | 0.28 | 0.32 (48.3) | 0.67; 0.15–2.51 | 0.41 | 0.24 (40.4) |
| Mg (%) | 0.33 | 0.11 (51.7) | 18 | 0.15 | 0.13 (57.8) | 0.23; 0.03–1.43 | 0.27 | 0.11 (48.6) |
| B (µg·g−1) | 0.35 | 10.47 (45.4) | 17 | 0.13 | 12.20 (52.9) | 24.3; 2.1–200.7 | 0.23 | 20.45 (96.4) |
| Fe (µg·g−1) | 0.26 | 15.96 (43.4) | 23 | 0.13 | 17.34 (47.2) | 37.5; 12.5–189.1 | 0.25 | 20.56 (61.8) |
| Structure and Defense | ||||||||
| C (%) | 0.61 | 1.81 (3.7) | 13 | 0.48 | 2.08 (4.2) | 49.6; 40.1–55.8 | 0.41 | 2.34 (4.7) |
| Lignin (%) | 0.45 | 7.59 (28.3) | 14 | 0.16 | 9.18 (34.3) | 26.8; 0.9–67.2 | 0.22 | 8.73 (32.4) |
| Cellulose (%) | 0.33 | 4.80 (28.2) | 16 | 0.06 | 5.64 (33.1) | 16.9; 1.1–36.9 | 0.18 | 5.34 (32.3) |
| Tannins (mg·g−1) * | 0.56 | 18.02 (25.9) | 19 | 0.22 | 23.30 (33.5) | 69.5; 12.8–149.1 | 0.39 | 24.62 (35.4) |
| Phenols (mg·g−1) * | 0.39 | 32.83 (26.9) | 21 | 0.04 | 41.36 (33.9) | 120.9; 22.6–240.2 | 0.18 | 44.02 (36.7) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, R.E.; Chadwick, K.D.; Brodrick, P.G.; Carranza-Jimenez, L.; Vaughn, N.R.; Asner, G.P. An Approach for Foliar Trait Retrieval from Airborne Imaging Spectroscopy of Tropical Forests. Remote Sens. 2018, 10, 199. https://doi.org/10.3390/rs10020199
Martin RE, Chadwick KD, Brodrick PG, Carranza-Jimenez L, Vaughn NR, Asner GP. An Approach for Foliar Trait Retrieval from Airborne Imaging Spectroscopy of Tropical Forests. Remote Sensing. 2018; 10(2):199. https://doi.org/10.3390/rs10020199
Chicago/Turabian StyleMartin, Roberta E., K. Dana Chadwick, Philip G. Brodrick, Loreli Carranza-Jimenez, Nicholas R. Vaughn, and Gregory P. Asner. 2018. "An Approach for Foliar Trait Retrieval from Airborne Imaging Spectroscopy of Tropical Forests" Remote Sensing 10, no. 2: 199. https://doi.org/10.3390/rs10020199
APA StyleMartin, R. E., Chadwick, K. D., Brodrick, P. G., Carranza-Jimenez, L., Vaughn, N. R., & Asner, G. P. (2018). An Approach for Foliar Trait Retrieval from Airborne Imaging Spectroscopy of Tropical Forests. Remote Sensing, 10(2), 199. https://doi.org/10.3390/rs10020199