Predictive Ecosystem Mapping of South-Eastern Australian Temperate Forests Using Lidar-Derived Structural Profiles and Species Distribution Models
Abstract
:1. Introduction
2. Materials and Methods
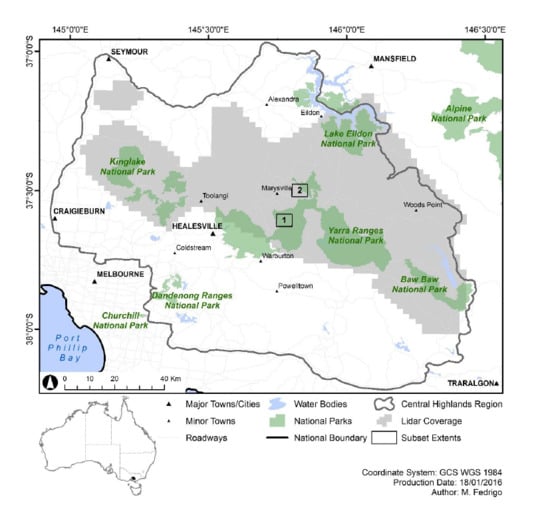
2.1. Study Area
2.2. Species Composition of Stand Types
2.3. Floristic Sampling
2.4. Lidar-Derived Digital Elevation Model (DEM)
2.5. Environmental and Satellite Data
2.6. Species Distribution Modelling
2.7. Predictive Ecosystem Mapping Model
3. Results
3.1. Species Composition of Stand Types
3.2. Species Distribution Modelling
3.3. Predictive Ecosystem Mapping and Comparison With Ecological Vegetation Classes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Franklin, J. Predictive vegetation mapping: Geographic modeling of biospatial patterns in relation to environmental gradients. Prog. Phys. Geog. 1995, 19, 474–499. [Google Scholar] [CrossRef]
- Miller, J. Species distribution modeling. Geogr. Compass 2010, 4, 490–509. [Google Scholar] [CrossRef]
- Cord, A.F.; Meentemeyer, R.K.; Leitão, P.J.; Václavík, T. Modelling species distributions with remote sensing data: Bridging disciplinary perspectives. J. Biogeogr. 2013, 40, 2226–2227. [Google Scholar] [CrossRef]
- Jones, K.; Meidinger, D.; Clark, D.; Schultz, F. Towards the Establishment of Predictive Ecosystem Mapping Standards: A White Paper; 1st Approximation. Prepared for Terrestrial Ecosystem Mapping Alternatives Task Force; Resource Inventory Committee (RIC), Victoria, BC, Canada, 8–9 September 1999; pp. 1–88. [Google Scholar]
- Meidinger, D.; Enns, B.; Banner, A.; Jones, C. EcoGen: A model for predictive ecosystem mapping. In Proceedings from Science to Management and Back: A Science Forum for Southern Interior Ecosystems of British Columbia; Hollstedt, C., Sutherland, K., Innes, T., Eds.; Southern Interior Forest Extension and Research Partnership: Kamloops, BC, Canada, 2000; pp. 45–47. [Google Scholar]
- Ohmann, J.L.; Gregory, M.J. Predictive mapping of forest composition and structure with direct gradient analysis and nearest-neighbor imputation in coastal Oregon, U.S.A. Can. J. For. Res. 2002, 32, 725–741. [Google Scholar] [CrossRef]
- Fraser, R.; McLennan, D.; Ponomarenko, S.; Olthof, I. Image-based predictive ecosystem mapping in Canadian arctic parks. Int. J. Appl. Earth Obs. 2012, 14, 129–138. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Kearney, M.; Porter, W. Mechanistic niche modelling: Combining physiological and spatial data to predict species’ ranges. Ecol. Lett. 2009, 12, 334–350. [Google Scholar] [CrossRef]
- Nix, H.A.; Busby, J. BIOCLIM, a Bioclimatic Analysis and Prediction System. Annual Report CSIRO; CSIRO Division of Water and Land Resources: Canberra, Australia, 1986.
- Pearson, R.G.; Dawson, T.P. Predicting the impacts of climate change on the distribution of species: Are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef]
- Zimble, D.A.; Evans, D.L.; Carlson, G.C.; Parker, R.C.; Grado, S.C.; Gerard, P.D. Characterizing vertical forest structure using small-footprint airborne LiDAR. Remote Sens. Environ. 2003, 87, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Hirzel, A.H.; Hausser, J.; Chessel, D.; Perrin, N. Ecological-niche factor analysis: How to compute habitat-suitability maps without absence data? Ecology 2002, 83, 2027–2036. [Google Scholar] [CrossRef]
- Vierling, K.T.; Vierling, L.A.; Gould, W.A.; Martinuzzi, S.; Clawges, R.M. Lidar: Shedding new light on habitat characterization and modeling. Front. Ecol. Environ. 2008, 6, 90–98. [Google Scholar] [CrossRef]
- Peterson, A.T. Uses and requirements of ecological niche models and related distributional models. Biodivers. Inform. 2006, 3, 59–72. [Google Scholar] [CrossRef]
- Stockwell, D.R.B. Improving ecological niche models by data mining large environmental datasets for surrogate models. Ecol. Model. 2006, 192, 188–196. [Google Scholar] [CrossRef] [Green Version]
- Booth, T.H. A new method for assisting species selection. Commonw. For. Rev. 1985, 64, 241–250. [Google Scholar]
- Austin, M.P. Spatial prediction of species distribution: An interface between ecological theory and statistical modelling. Ecol. Model. 2002, 157, 101–118. [Google Scholar] [CrossRef]
- Ferrier, S.; Drielsma, M.; Manion, G.; Watson, G. Extended statistical approaches to modelling spatial pattern in biodiversity in north-east New South Wales. II. Community-level modelling. Biodivers. Conserv. 2002, 11, 2309–2338. [Google Scholar] [CrossRef]
- Overton, J.M.; Stephens, R.T.T.; Leathwick, J.R.; Lehmann, A. Information pyramids for informed biodiversity conservation. Biodivers. Conserv. 2002, 11, 2093–2116. [Google Scholar] [CrossRef]
- Ferrier, S.; Guisan, A. Spatial modelling of biodiversity at the community level. J. Appl. Ecol. 2006, 43, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J.; Gordon, A.; Kujala, H.; Lentini, P.E.; McCarthy, M.A.; Tingley, R.; Wintle, B.A. Is my species distribution model fit for purpose? Matching data and models to applications. Glob. Ecol. Biogeogr. 2015, 24, 276–292. [Google Scholar] [CrossRef] [Green Version]
- Maguire, K.C.; Nieto-Lugilde, D.; Fitzpatrick, M.C.; Williams, J.W.; Blois, J.L. Modeling species and community responses to past, present, and future episodes of climatic and ecological change. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 343–368. [Google Scholar] [CrossRef]
- Ohmann, J.L.; Gregory, M.J.; Henderson, E.B.; Roberts, H.M. Mapping gradients of community composition with nearest-neighbour imputation: Extending plot data for landscape analysis. J. Veg. Sci. 2011, 22, 660–676. [Google Scholar] [CrossRef]
- Clark, J.S.; Gelfand, A.E.; Woodall, C.W.; Zhu, K. More than the sum of the parts: Forest climate response from joint species distribution models. Ecol. Appl. 2014, 24, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Pollock, L.J.; Tingley, R.; Morris, W.K.; Golding, N.; O’Hara, R.B.; Parris, K.M.; Vesk, P.A.; McCarthy, M.A. Understanding co-occurrence by modelling species simultaneously with a Joint Species Distribution Model (JSDM). Methods Ecol. Evol. 2014, 5, 397–406. [Google Scholar] [CrossRef] [Green Version]
- Cutler, D.R.; Edwards, T.C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef] [PubMed]
- Olden, J.D.; Lawler, J.J.; Poff, N.L. Machine learning methods without tears: A primer for ecologists. Q. Rev. Biol. 2008, 83, 171–193. [Google Scholar] [CrossRef] [PubMed]
- Hakkenberg, C.R.; Peet, R.K.; Urban, D.L.; Song, C. Modeling plant composition as community continua in a forest landscape with LiDAR and hyperspectral remote sensing. Ecol. Appl. 2018, 28, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Recknagel, F. Ecological Informatics: Scope, Techniques and Applications, 2nd ed.; Springer: New York, NY, USA, 2006; pp. 1–496. ISBN 9783540283836. [Google Scholar]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- De’ath, G. Boosted trees for ecological modeling and prediction. Ecology 2007, 88, 243–251. [Google Scholar] [CrossRef]
- Wilson, B.T.; Lister, A.J.; Riemann, R.I. A nearest-neighbor imputation approach to mapping tree species over large areas using forest inventory plots and moderate resolution raster data. For. Ecol. Manag. 2012, 271, 182–198. [Google Scholar] [CrossRef]
- Helmer, E.; Ruzycki, T.; Wilson, B.; Sherrill, K.; Lefsky, M.; Marcano-Vega, H.; Brandeis, T.; Erickson, H.; Ruefenacht, B. Tropical deforestation and recolonization by exotic and native trees: Spatial patterns of tropical forest biomass, functional groups, and species counts and links to stand age, geoclimate, and sustainability goals. Remote Sens. 2018, 10, 1724. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N.E. Predictive habitat distribution models in ecology. Ecol. Model. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Araújo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Heikkinen, R.K.; Luoto, M.; Araújo, M.B.; Virkkala, R.; Thuiller, W.; Sykes, M.T. Methods and uncertainties in bioclimatic envelope modelling under climate change. Prog. Phys. Geogr. 2006, 30, 751–777. [Google Scholar] [CrossRef] [Green Version]
- Austin, M.P.; Van Neil, K.P. Improving species distribution models for climate change studies: Variable selection and scale. J. Biogeogr. 2011, 38, 1–8. [Google Scholar] [CrossRef]
- Morrissey, R.C.; Jacobs, D.F.; Seifert, J.R.; Kershaw, J.A. Overstory species composition of naturally regenerated clearcuts in an ecological classification framework. Plant Ecol. 2010, 208, 21–34. [Google Scholar] [CrossRef]
- McElhinny, C.; Gibbons, P.; Brack, C.; Bauhus, J. Forest and woodland stand structural complexity: Its definition and measurement. For. Ecol. Manag. 2005, 218, 1–24. [Google Scholar] [CrossRef]
- Wilkes, P.; Jones, S.D.; Suarez, L.; Heywood, A.; Mellor, A.; Woodgate, W.; Soto-Berelov, M.; Skidmore, A. Using discrete-return ALS to quantify number of canopy strata across diverse forest types. Methods Ecol. Evol. 2016, 7, 700–712. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ole, H.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Lefsky, M.A.; Cohen, W.B.; Parker, G.G.; Harding, D.J. Lidar remote sensing for ecosystem studies. BioScience 2002, 52, 19–30. [Google Scholar] [CrossRef]
- Brubaker, K.M.; Johnson, Q.K.; Kaye, M.W. Spatial patterns of tree and shrub biomass in a deciduous forest using leaf-off and leaf-on lidar. Can. J. For. Res. 2018, 48, 1020–1033. [Google Scholar] [CrossRef]
- Hopkinson, C.; Chasmer, L.; Young-Pow, C.; Treitz, P. Assessing forest metrics with a ground-based scanning lidar. Can. J. For. Res. 2004, 34, 573–583. [Google Scholar] [CrossRef]
- Bergen, K.M.; Goetz, S.J.; Dubayah, R.O.; Henebry, G.M.; Hunsaker, C.T.; Imhoff, M.L.; Nelson, R.F.; Parker, G.G.; Radeloff, V.C. Remote sensing of vegetation 3-D structure for biodiversity and habitat: Review and implications for lidar and radar spaceborne missions. J. Geophys. Res. Biogeogr. 2009, 114, 1–13. [Google Scholar] [CrossRef]
- Pirotti, F. Analysis of full-waveform LiDAR data for forestry applications: A review of investigations and methods. iForest-Biogeosci. For. 2011, 4, 100–106. [Google Scholar] [CrossRef]
- Marselis, S.M.; Tang, H.; Armston, J.D.; Calders, K.; Labrière, N.; Dubayah, R. Distinguishing vegetation types with airborne waveform lidar data in tropical forest-savanna mosaic: A case study in Lopé National Park, Gabon. Remote Sens. Environ. 2018, 216, 626–634. [Google Scholar] [CrossRef]
- Kane, V.R.; McGaughey, R.J.; Bakker, J.D.; Gersonde, R.F.; Lutz, J.A.; Franklin, J.F. Comparisons between field-and LiDAR-based measures of stand structural complexity. Can. J. For. Res. 2010, 40, 761–773. [Google Scholar] [CrossRef]
- Simonson, W.D.; Allen, H.D.; Coomes, D.A. Use of an airborne lidar system to model plant species composition and diversity of Mediterranean oak forests. Conserv. Biol. 2012, 26, 840–850. [Google Scholar] [CrossRef]
- Broadbent, E.N.; Asner, G.P.; Peña-Claros, M.; Palace, M.; Soriano, M. Spatial partitioning of biomass and diversity in a lowland Bolivian forest: Linking field and remote sensing measurements. For. Ecol. Manag. 2008, 255, 2602–2616. [Google Scholar] [CrossRef]
- Liu, X. Airborne LiDAR for DEM generation: Some critical issues. Prog. Phys. Geogr. 2008, 32, 31–49. [Google Scholar] [CrossRef]
- Campbell, M.J.; Dennison, P.E.; Hudak, A.T.; Parham, L.M.; Butler, B.W. Quantifying understory vegetation density using small-footprint airborne lidar. Remote Sen. Environ. 2018, 215, 330–342. [Google Scholar] [CrossRef]
- Moran, C.J.; Rowell, E.M.; Seielstad, C.A. A data-driven framework to identify and compare forest structure classes using LiDAR. Remote Sens. Environ. 2018, 211, 154–166. [Google Scholar] [CrossRef]
- Dubayah, R.O.; Drake, J.B. Lidar remote sensing for forestry. J. For. 2000, 98, 44–46. [Google Scholar] [CrossRef]
- Lim, K.; Treitz, P.; Wulder, M.; St-Onge, B.; Flood, M. LiDAR remote sensing of forest structure. Prog. Phys. Geogr. Earth Environ. 2003, 27, 88–106. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Evans, J.S.; Martinuzzi, S.; Gessler, P.E.; Hudak, A.T. Characterizing forest succession with lidar data: An evaluation for the inland northwest, USA. Remote Sens. Environ. 2009, 113, 946–956. [Google Scholar] [CrossRef]
- Van Leeuwen, M.; Nieuwenhuis, M. Retrieval of forest structural parameters using LiDAR remote sensing. Eur. J. For. Res. 2010, 129, 749–770. [Google Scholar] [CrossRef]
- Hakkenberg, C.R.; Zhu, K.; Peet, R.K.; Song, C. Mapping multi-scale vascular plant richness in a forest landscape with integrated LiDAR and hyperspectral remote-sensing. Ecology 2018, 99, 474–487. [Google Scholar] [CrossRef]
- Fedrigo, M.; Newnham, G.J.; Coops, N.C.; Culvenor, D.S.; Bolton, D.K.; Nitschke, C.R. Predicting temperate forest stand types using only structural profiles from discrete return airborne lidar. ISPRS J. Photogramm. 2018, 136, 106–119. [Google Scholar] [CrossRef]
- Cameron, D. A Field Guide to Rainforest Identification in Victoria: Differential Species Keys for the Delineation of Rainforest Boundaries; Department of Sustainability and Environment: Melbourne, Australia, 2011; ISBN 9781742871219.
- Kasel, S.; Bennett, L.T.; Aponte, C.; Fedrigo, M.; Nitschke, C.R. Environmental heterogeneity promotes floristic turnover in temperate forests of south-eastern Australia more than dispersal limitation and disturbance. Landsc. Ecol. 2017, 32, 1613–1629. [Google Scholar] [CrossRef]
- Muir, A.M.; Edwards, S.A.; Dickins, M.J. Description and Conservation Status of the Vegetation of the Box-Ironbark Ecosystem in Victoria; Department of Conservation and Natural Resources: Victoria, Australia, 1995.
- Department of Sustainability and Environment (DSE), Victorian Government Department of Sustainability and Environment Melbourne. Native Vegetation Information: Native Vegetation Extent Dataset Information Sheet No 3. 2007. Available online: http://www.depi.vic.gov.au/__data/assets/pdf_file/0008/97325/NV_extent_dataset.pdf (accessed on 17 July 2015).
- Fedrigo, M.; Kasel, S.; Bennett, L.T.; Roxburgh, S.H.; Nitschke, C.R. Carbon stocks in temperate forests of south-eastern Australia reflect large tree distribution and edaphic conditions. For. Ecol. Manag. 2014, 334, 129–143. [Google Scholar] [CrossRef]
- Woodgate, P.W.; Peel, B.D.; Coram, J.E.; Farrell, S.J.; Ritman, K.T.; Lewis, A. Old-growth forest studies in Victoria, Australia concepts and principles. For. Ecol. Manag. 1996, 85, 79–94. [Google Scholar] [CrossRef]
- Department of Environment, Land, Water and Planning (DELWP), Victoria State Government. EVC Benchmarks. 2015. Available online: http://www.depi.vic.gov.au/environment-and-wildlife/biodiversity/evc-benchmarks#bioregionname (accessed on 17 July 2015).
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Method; PRIMER-E Ltd.: Plymouth Marine Laboratory, UK, 2008. [Google Scholar]
- Clarke, K.R.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation; PRIMER-E Ltd.: Plymouth Marine Laboratory, UK, 2001; ISBN 9781855311404. [Google Scholar]
- Boland, D.J.; Brooker, M.I.H.; Turnbull, J.W. Eucalyptus Seed; CSIRO: Canberra, Australia, 1980; pp. 1–191.
- Close, D.C.; Wilson, S.J. Provenance effects on pre-germination treatments for Eucalyptus regnans and E. delegatensis seed. For. Ecol. Manag. 2002, 170, 299–305. [Google Scholar] [CrossRef]
- Fedrigo, M.; Stewart, S.B.; Kasel, S.; Levchenko, V.; Trouvé, R.; Nitschke, C.R. Radiocarbon dating informs tree fern population dynamics and disturbance history of temperate forests in southeast Australia. Radiocarbon 2018, in press. [Google Scholar] [CrossRef]
- Vierling, K.T.; Swift, C.E.; Hudak, A.T.; Vogeler, J.C.; Vierling, L.A. How much does the time lag between wildlife field-data collection and LiDAR-data acquisition matter for studies of animal distributions? A case study using bird communities. Remote Sens. Lett. 2014, 5, 185–193. [Google Scholar] [CrossRef]
- Scarth, P.; Röder, A.; Schmidt, M. Tracking Grazing Pressure and Climate Interaction—The Role of Landsat Fractional Cover in Time Series Analysis. In Proceedings of the 15th Australasian Remote Sensing and Photogrammetry Conference (ARSPC), Alice Springs, Australia, 13–17 September 2010. [Google Scholar]
- Flood, N. Seasonal composite landsat TM/ETM+ images using the Medoid (a multi-dimensional median). Remote Sens. 2013, 5, 6481–6500. [Google Scholar] [CrossRef]
- Seasonal Fractional Cover—Landsat, JRSRP Algorithm, Australia Coverage. Joint Remote Sensing Research Program (JRSRP). 2016. Available online: http://auscover.org.au/purl/landsat-seasonal-fractional-cover (accessed on 20 December 2016).
- Pettorelli, N.; Wegmann, M.; Skidmore, A.; Mücher, S.; Dawson, T.P.; Fernandez, M.; Lucas, R.; Schaepman, M.E.; Wang, T.; O’Connor, B.; et al. Framing the concept of satellite remote sensing essential biodiversity variables: Challenges and future directions. Remote Sens. Ecol. Conserv. 2016, 2, 122–131. [Google Scholar] [CrossRef]
- Conrad, O.; Bechel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for automated geoscientific analyses (SAGA) v. 2.1.4. Geosci. Model Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- McCune, B.; Mefford, M.J. HyperNiche. Nonparametric Multiplicative Habitat Modeling, version 2; MjM Software: Gleneden Beach, OR, USA, 2009. [Google Scholar]
- Wang, T.; Hamann, A.; Spittlehouse, D.L.; Aitken, S.N. Development of scale-free climate data for western Canada for use in resource management. Int. J. Climatol. 2006, 26, 383–397. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Phillips, S.; Leathwick, J.; Elith, J. dismo: Species distribution modeling. R package version 1.1-4. 2017. Available online: https://CRAN.R-project.org/package=dismo (accessed on 12 January 2017).
- R: A Language and Environment for Statistical Computing; R Core Team, R Foundation for Statistical Computing: Vienna, Austria, 2016. Available online: https://www.R-project.org/ (accessed on 1 January 2013).
- Stewart, S.B.; Nitschke, C.R. Improving temperature interpolation using MODIS LST and local topography: A comparison of methods in south east Australia. Int. J. Climatol. 2017, 37, 3098–3110. [Google Scholar] [CrossRef]
- Hutchinson, M.F.; Xu, T. ANUSPLIN Version 4.4 User Guide; Fenner School of Environment and Society, Australian National University: Canberra, Australia, 2013. [Google Scholar]
- Fenner School of Environment and Society and Geoscience Australia. GEODATA 9 Second Digital Elevation Model (DEM-9S), Version 3; Fenner School of Environment and Society and Geoscience Australia: Canberra, Australia, 2008. [Google Scholar]
- Stewart, S.B.; Choden, K.; Fedrigo, M.; Roxburgh, S.H.; Keenan, R.; Nitschke, C.R. The role of topography and the north Indian monsoon on mean monthly climate interpolation within the Himalayan Kingdom of Bhutan. Int. J. Climatol. 2017, 37 (Suppl. 1), 897–909. [Google Scholar] [CrossRef]
- Hopkinson, R.F.; Hutchinson, M.F.; McKenney, D.W.; Milewska, E.J.; Padadopol, P. Optimizing input data for gridding climate normals for Canada. J. Appl. Meteorol. Clim. 2012, 51, 1508–1518. [Google Scholar] [CrossRef]
- Alduchov, O.A.; Eskridge, R.E. Improved Magnus form approximation of saturation vapour pressure. J. Appl. Meteorol. 1996, 34, 601–609. [Google Scholar] [CrossRef]
- Lawrence, M. The relationship between relative humidity and the dewpoint temperature in moist air: A simple conversion and applications. Bull. Am. Meteorol. Soc. 2005, 86, 225–233. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; García Marquéz, J.R.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- O’Donnell, M.S.; Ignizio, D.A. Bioclimatic predictors for supporting ecological applications in the conterminous United States. US Geol. Surv. Data Ser. 2012, 691, 1–10. [Google Scholar]
- Hutchinson, M.F. Interpolation of rainfall data with thin plate smoothing splines—Part II: Analysis of topographic dependence. GIDA 1998, 2, 152–167. [Google Scholar]
- Sharples, J.J.; Hutchinson, M.F.; Jellett, D.R. On the horizontal scale of elevation dependence of Australian monthly precipitation. J. Appl. Meteorol. 2005, 44, 1850–1865. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer classification and regression tree techniques: Bagging and random forests for ecological prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Department of Environment, Land, Water and Planning (DELWP), Victoria State Government. Victorian Biodiversity Atlas. 2015. Available online: https://vba.dse.vic.gov.au/vba/ (accessed on 10 April 2015).
- Valavi, R.; Elith, J.; Lahoz-Monfort, J.J.; Guillera-Arroita, G. blockCV: An R package for generating spatially or environmentally separated folds for k-fold cross-validation of species distribution models. Methods Ecol. Evol. 2018. [Google Scholar] [CrossRef]
- Jiménez-Valverde, A.; Lobo, J.M. Threshold criteria for conversion of probability of species presence to either-or presence-absence. Acta Oecol. 2007, 31, 361–369. [Google Scholar] [CrossRef]
- Ferrier, S.; Manion, G.; Elith, J.; Richardson, K. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Divers. Distrib. 2007, 13, 252–264. [Google Scholar] [CrossRef]







| Lidar System Configurations | |
|---|---|
| Acquisition date range | 19 November 2007–0 January 2008 |
| Sensor type | OptechALTM3100EA |
| Wavelength (nm) | 1064 |
| Scan rate (kHz) | 71 |
| Scan angle (°) | ±25 |
| Mean footprint size (m) | 0.26 |
| Pulses (m2) | 0.90 |
| Maximum returned signals | 4 |
| Horizontal accuracy (cm) | ±35 |
| Vertical accuracy (cm) | ±50 |
| Variable | Minimum | Maximum | Mean |
|---|---|---|---|
| Environmental—Topography | |||
| Elevation (m above sea level) | 103.9 | 1567.9 | 652.0 |
| SlopeA (°) | 0 | 68.1 | 17.1 |
| Topographic Position IndexA (TPI; 100 m radius) | −8.6 | 14.2 | 0 |
| Heat load indexB (HLI) | 0.2 | 1.0 | 0.8 |
| Potential direct incident radiationB (PDIR; MJ cm−2 yr−1) | 0.2 | 1.0 | 0.8 |
| Proximity to waterwaysC (m) | 0.0 | 6882.7 | 1817.5 |
| Environmental - Climate | |||
| Annual mean temperature (BIO1; °C) | 5.9 | 14.9 | 11.6 |
| Mean diurnal range (BIO2; °C) | 5.1 | 13.4 | 9.2 |
| Isothermality (BIO3; %) | 28.8 | 51.8 | 43.4 |
| Temperature seasonality (BIO4; standard deviation; °C) | 3.7 | 5.0 | 4.3 |
| Maximum temperature of warmest month (BIO5; °C) | 16.2 | 29.1 | 23.8 |
| Minimum temperature of coldest month (BIO6; °C) | −2.4 | 5.7 | 2.6 |
| Temperature annual range (BIO7; °C) | 16.7 | 27.0 | 21.2 |
| Mean temperature of wettest quarter (BIO8; °C) | 1.6 | 13.5 | 6.8 |
| Mean temperature of driest quarter (BIO9; °C) | 11.1 | 19.8 | 16.7 |
| Mean temperature of warmest quarter (BIO10; °C) | 11.1 | 20.8 | 16.9 |
| Mean temperature of coldest quarter (BIO11; °C) | 0.7 | 9.5 | 6.3 |
| Annual mean precipitation (BIO12; mm) | 667.1 | 2089.5 | 1290.3 |
| Precipitation of wettest month (BIO13; mm) | 70.7 | 274.9 | 164.9 |
| Precipitation of driest month (BIO14; mm) | 31.8 | 84.2 | 53.0 |
| Precipitation seasonality (BIO15; coefficient of variation; %) | 14.8 | 47.2 | 31.9 |
| Precipitation of wettest quarter (BIO16; mm) | 202.3 | 745.4 | 454.3 |
| Precipitation of driest quarter (BIO17; mm) | 109.7 | 324.3 | 194.7 |
| Precipitation of warmest quarter (BIO18; mm) | 110.5 | 324.3 | 198.5 |
| Precipitation of coldest quarter (BIO19; mm) | 199.5 | 690.0 | 446.6 |
| Annual mean vapour pressure deficit (hPa) | 2.0 | 6.2 | 4.0 |
| Summer mean vapour pressure deficit (hPa) | 3.6 | 10.7 | 6.9 |
| Annual heat moisture indexD (AHMI) | 7.7 | 35.9 | 17.8 |
| Satellite | |||
| Bare ground fraction (% + 100) | 97.7 | 194.1 | 103.5 |
| Green vegetation fraction (% + 100) | 98.0 | 203.1 | 169.0 |
| Non-green vegetation fraction (% + 100) | 95.9 | 202.0 | 126.0 |
| Species | Predictors | P | A | T | TPR | TNR | OA | TSS | AUC |
|---|---|---|---|---|---|---|---|---|---|
| Atherosperma moschatum | Environmental | 38 | 423 | 0.22 | 0.84 | 0.80 | 0.80 | 0.64 | 0.87 |
| Environmental and Satellite | 38 | 423 | 0.14 | 0.82 | 0.78 | 0.79 | 0.60 | 0.83 | |
| Eucalyptus delegatensis | Environmental | 57 | 344 | 0.24 | 0.98 | 0.81 | 0.84 | 0.79 | 0.92 |
| Environmental and Satellite | 57 | 344 | 0.41 | 0.96 | 0.88 | 0.89 | 0.84 | 0.92 | |
| Eucalyptus regnans | Environmental | 92 | 273 | 0.25 | 0.86 | 0.73 | 0.76 | 0.58 | 0.81 |
| Environmental and Satellite | 92 | 273 | 0.20 | 0.99 | 0.72 | 0.79 | 0.71 | 0.85 | |
| Nothofagus cunninghamii | Environmental | 129 | 365 | 0.15 | 0.95 | 0.64 | 0.72 | 0.59 | 0.83 |
| Environmental and Satellite | 129 | 365 | 0.21 | 0.90 | 0.68 | 0.74 | 0.58 | 0.84 | |
| Pittosporum bicolor | Environmental | 52 | 264 | 0.16 | 0.88 | 0.66 | 0.70 | 0.54 | 0.81 |
| Environmental and Satellite | 52 | 264 | 0.19 | 0.87 | 0.72 | 0.75 | 0.59 | 0.80 |
| Stand Type | Extent 1 | Extent 2 | Landscape | |||
|---|---|---|---|---|---|---|
| Species Model | Structure Model | Species Model | Structure Model | Species Model | Structure Model | |
| Rainforest | 7.8 (107.6) | 4.1 (10.2) | 7.5 (267.5) | 2.3 (15.3) | 783.51 (509.2) | 219.45 (70.6) |
| Ecotone | 11.5 (36.0) | 12.4 (45.6) | 11.5 (−6.3) | 15.7 (27.8) | 477.30 (−26.9) | 2921.27 (347.4) |
| Eucalypt | 5.7 (−11.5) | 8.5 (32.3) | 6.0 (−20.2) | 6.2 (−17.4) | 3911.82 (234.5) | 1712.16 (46.4) |
| Stand Type | Extent 1 | Extent 2 | Landscape | ||||
|---|---|---|---|---|---|---|---|
| PEM | EVC | PEM | EVC | PEM | EVC | PEM | EVC |
| Rainforest | Cool Temperate Rainforest | 3.7 | 5.4 | 2.0 | 4.5 | 128.6 (2.5) | 101.8 (2.0) |
| Ecotone | Not classified (NC) | 8.5 | NC | 12.3 | NC | 652.9 (12.6) | NC |
| Eucalypt | Wet Forest | 6.4 | 8.8 | 7.6 | 13.1 | 1169.4 (22.6) | 717.5 (13.9) |
| Damp Forest | - | 0.0 | - | 0.0 | - | 970.9 (18.8) | |
| Shrubby Wet Forest | - | 0.0 | - | 0.0 | - | <0.1 (0.0) | |
| Riparian | Riparian Forest | 0.4 | 0.0 | 0.3 | 0.0 | 90.8 (1.8) | 204.3 (3.9) |
| Montane Eucalypt | Montane Wet Forest | 5.3 | 10.8 | 2.0 | 7.4 | 132.9 (2.6) | 458.0 (8.8) |
| Montane Damp Forest | - | 0.0 | - | 0.0 | - | 211.5 (4.1) | |
| Sub-Alpine Woodland | Sub-Alpine Woodland | 0.6 | 0.0 | 0.0 | 0.0 | 90.6 (1.7) | 81.3 (1.6) |
| Other Forest | All Other Vegetation Classes | 0.0 | 0.0 | 0.1 | 0.0 | 2586.6 (50.0) | 2135.0 (41.3) |
| Non-Forest | Non-Forest | 0.0 | 0.0 | 0.7 | 0.0 | 323.1 (6.2) | 294.5 (5.7) |
| Ecological Vegetation Class | Extent 1 | Extent 2 | Landscape |
|---|---|---|---|
| Cool Temperate Rainforest | 2.2 (25.8) | 2.2 (17.9) | 42.8 (6.5) |
| Wet Forest | 3.6 (43.0) | 7.4 (60.6) | 230.6 (35.3) |
| Damp Forest | 0.0 | 0.0 | 90.5 (13.9) |
| Shrubby Wet forest | 0.0 | 0.0 | 0.0 |
| Riparian Forest | 0.0 | 0.0 | 31.1 (4.8) |
| Montane Wet Forest | 2.6 (31.2) | 2.6 (21.4) | 172.1 (26.4) |
| Montane Damp Forest | 0.0 | 0.0 | 33.8 (5.2) |
| Sub-Alpine Woodland | 0.0 | 0.0 | 9.0 (1.4) |
| All Other Vegetation Classes | 0.0 | 0.0 | 41.9 (6.4) |
| Non-Forest | 0.0 | 0.0 | 1.0 (0.1) |
| Total Ecotone Area | 8.5 | 12.3 | 652.9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedrigo, M.; Stewart, S.B.; Roxburgh, S.H.; Kasel, S.; Bennett, L.T.; Vickers, H.; Nitschke, C.R. Predictive Ecosystem Mapping of South-Eastern Australian Temperate Forests Using Lidar-Derived Structural Profiles and Species Distribution Models. Remote Sens. 2019, 11, 93. https://doi.org/10.3390/rs11010093
Fedrigo M, Stewart SB, Roxburgh SH, Kasel S, Bennett LT, Vickers H, Nitschke CR. Predictive Ecosystem Mapping of South-Eastern Australian Temperate Forests Using Lidar-Derived Structural Profiles and Species Distribution Models. Remote Sensing. 2019; 11(1):93. https://doi.org/10.3390/rs11010093
Chicago/Turabian StyleFedrigo, Melissa, Stephen B. Stewart, Stephen H. Roxburgh, Sabine Kasel, Lauren T. Bennett, Helen Vickers, and Craig R. Nitschke. 2019. "Predictive Ecosystem Mapping of South-Eastern Australian Temperate Forests Using Lidar-Derived Structural Profiles and Species Distribution Models" Remote Sensing 11, no. 1: 93. https://doi.org/10.3390/rs11010093
APA StyleFedrigo, M., Stewart, S. B., Roxburgh, S. H., Kasel, S., Bennett, L. T., Vickers, H., & Nitschke, C. R. (2019). Predictive Ecosystem Mapping of South-Eastern Australian Temperate Forests Using Lidar-Derived Structural Profiles and Species Distribution Models. Remote Sensing, 11(1), 93. https://doi.org/10.3390/rs11010093