1. Introduction
Precision Agriculture or Smart Farming aims to increase crop yield, reduce production costs and decrease environmental impact. In this context, an active research area is to identify crops automatically in digital images to classify plants, to monitor its growth or to detect problems of water stress, nutrition or health in cultivated plants. This problem is complicated under open field cultivation due to different factors such as natural lighting, weather and agricultural practices of the farmers.
The research carried out so far has been limited only to cases where the open field crops have small plants that are well separated from one another, the colour of the soil with respect to the plants is very different and the overlap among leaves of the same plant occurs very rarely [
1,
2,
3]. Moreover, the state-of-the-art has focused on annual crops with careful cultivation techniques, without addressing the accurate segmentation case of fig (
Ficus carica) perennial plants in an orchard where their particular characteristics [
4] cause complex image patterns.
Leaves and fruits of fig have several nutritional and medicinal properties and currently the interest in its production has increased worldwide [
5]. According to FAOSTAT report 2016, the harvested area of fig in the world was estimated around 308,460 hectares with a production of 1,050,459 tons [
6]. The top five producers are Turkey, Egypt, Algeria, Iran and Morocco. In the case of México, the fig export market has been recently opened for the United States, which foresees an increase of fig planted area. In 2015, the cultivation area was about 1199 hectares yielding a total of 5381 tons with a value of approximately US
$ 2,839,500.00 [
7].
On the other hand, Unmanned Aerial Vehicles (UAVs) have many characteristics that make them attractive elements for precision agriculture [
8]. UAVs can continuously travel large tracts of cultivated land in a short time and they have the capacity to carry light-weight compact sensors to capture information at low altitude. The RGB (Red-Green-Blue) cameras are one of the most used sensors in UAVs since they are relatively cheap, have low energy consumption and are light. It is true that multispectral or thermal cameras have been extensively used to vegetation monitoring [
9,
10], however these cameras are more expensive compared to RGB cameras.
In this paper, we address the problem of crop segmentation at pixel level. Our approach exploits relevant information from high resolution RGB images captured by an UAV in a difficult open field environment. Indeed, we are considering realistic environmental conditions where there are illumination variations and different types of soil and weed, most treetops are overlapping, the inter-row space of field crop is not constant and there may be various elements that are not of interest, for example, stones or objects used by farmers. Furthermore, we deal with the specific case of a crop of woody and tall fig plants, which leads to additional problems in top-view images. The branches of fig plants grow around and along the stem and their leaves are divided into 7 lobes.
Figure 1 shows examples of ground and aerial views of fig shrubs in which the plant morphology and the cultivation conditions are appreciated. Factors like camera position, solar illumination and plant morphology originate a visual appearance of the leaves that is drastically variable due to the formation of specularities, shadows, occlusions and different shapes, even though they have been captured at the same time in the morning.
We propose the use of a Convolutional Neural Network (CNN) with an encoder-decoder architecture trained end-to-end as the means to address the problem of plant segmentation at the granularity of pixel. Since artificial neural networks are highly robust approximation functions [
11], which when used with convolutional layers have set the state-of-the-art for dealing with different image-related tasks [
12,
13,
14], it is reasonable to expect that they could be used to perform segmentation in such a challenging scenario as ours. Furthermore, an encoder-decoder architecture provides the tools required to map RGB images onto binary images corresponding to segmentation indicator. Our model allows to classify each pixel of an image into
crop or
non-crop by using only the raw RGB pixel intensity values as input. In addition, we present an evaluation of algorithms based on the RGB colour model to detect vegetation, classical algorithms that have been considered as a reference for this type of analysis. We make available our CNN code and the data used for evaluation. To the best of our knowledge, this is the first public data set containing both high-resolution aerial images of tall fig shrubs under real open field cultivation conditions and hand-made ground truth segmentations, in contrast to previous work which are somehow limited, as they have focused only on small plants with little foliage grown in a controlled open field environment [
15]. The high resolution of the images allows to capture with greater detail the features of the plants, which is of great value for the resolution of the diverse problems that Precision Agriculture tries to solve. The code and data are released at:
https://github.com/jofuepa/fig-datasetThe contributions of this paper are as follows:
A CNN approach for accurate crop segmentation in a fig orchard using only RGB data, where the analysed plants grow under a great variability of circumstances, such as natural illumination and crop maintenance determined mainly by the experience of a small farmer. The proposed CNN has comparable performance with the state of the art and it can be trained in less time than SegNet-Basic [
14].
A public data set of high-resolution aerial images, captured by an RGB camera mounted on a UAV that flies at low altitude, of a field of figs of approximately one hectare and their corresponding Ground Truth (GT), where the leaves belonging to plants were labelled by hand with pixel level precision. The difficulty of segmentation in presented images are more challenging than the previous data sets because plants are not arranged along lines and each of the leaves of the plant occupy a very small region of the whole image.
The paper consists of the following sections.
Section 2 reviews the related work. The fig data set and the proposed network are introduced in
Section 3 and
Section 4, respectively.
Section 5 presents the experimental results. Finally, the conclusions and possible future work are provided in
Section 6.
2. Related Work
There has been a great success in the use of Deep Learning to solve a variety of problems in Speech Recognition, Computer Vision, Natural Language Understanding, Autonomous Driving and many others [
11,
12]. In the specific case of the Semantic Segmentation problem, whose objective is to categorize each pixel of an image, deep CNNs have shown to obtain better performance in large segmentation datasets of the state-of-the-art than traditional Machine Learning approaches [
13,
14,
16]. Despite these advances, CNNs and Semantic Segmentation principles are not yet widely adopted in agricultural tasks where it is possible to have digital images as data, for example, plant recognition, fruit counting and leaf classification [
17]. Kamilaris et al. [
15] signal that there existed only approximately 20 research efforts employing CNN to address various agricultural problems.
In the domain of plant recognition, Ye et al. [
18] examine the problem of corn crop detection in colour images under different intensities of natural lighting. The image acquisition is carried out by a camera placed on a post at a height of 5m. They propose a probabilistic Markov random field using superpixels and the neighbourhood relationships that exist between them. They treat the cases of leaves extraction that are under both the shadows and the white light spots produced by specular reflections in an environment where the crop is free of weeds. Li et al. [
19] perform cotton detection in a boll opening growth stage with a complicated background. Regions of pixels are extracted to perform a semantic segmentation by using a Random forest classifier. A problem with the aforementioned research is the need for superpixels creation and an image transformation of RGB to CIELAB colour space, which can be slow and imprecise.
There are approaches that focus mainly on carrying out a crop and weed segmentation with the objective of making a controlled application of herbicides. The use of robots equipped with cameras and other sensors has increased in order to perform this task. For instance, Milioto et al. [
20] propose a pixel-wise semantic segmentation of sugar beet plants, weeds and soil in colour images based on a CNN, dealing with natural lighting, soil and weather conditions. They capture the images using a ground robot and carry out an evaluation considering several phenological stages of the plant. However, this solution is tailored to deal with sugar beets, which is a biennial plant and its leaves can only reach a height of up to 0.35 m, while the cultivation of fig is perennial (30–40 years of life) and the plants are mostly 5–10 m high [
21]. Another method is presented in Reference [
22], the authors use a Fully CNN considering image sequences of sugar beet fields for crop and weed detection. They take into account 4-channel images (red, green, blue and near infra-red) and the spatial arrangement of row plants to obtain good pixel-wise semantic segmentation. Sa et al. [
10] analyse the crop/weed classification performance using dense semantic segmentation with different multispectral information as input to the SegNet network [
14]. Their images are collected by a micro aerial vehicle and a 4-band multispectral camera in a sugar beet crop. They conclude that the configuration of near infra-red, red channel and Normalized Difference Vegetation Index (NDVI) is the best for sugar beet detection.
In this paper, we focus on the accurate crop segmentation using only colour images as input information, weed or other elements are considered of little interest, with the aim of contributing in applications oriented to obtain reliable parameters of plant growth in an automatic way, for example, the leaf area index [
23]. The most studied crops in Precision Farming literature are plants that are short, have a short life cycle and are in a carefully cultivated state: carrot [
1], lettuce [
2], sugar beet [
3], maize [
24], cauliflower [
25] and radish [
26,
27]. However, these plants do not contain all the challenges that are present in fig plants growing in a complex environment where the crop maintenance is not carried out correctly.
One of the main drawbacks of applying supervised learning algorithms in Precision Farming is the lack of large public datasets with enough labelled images for training [
15]. To solve this problem, Milioto et al. [
20] use as input data for a CNN a total of 14 channels per image (raw RGB data, vegetation indexes, HSV colour channels and edge detectors) which allowed a better generalization for the problem despite the limited training data. Meanwhile, Di Cicco et al. [
28] generate a large synthetic dataset to train a model that detects sugar beet crops and weed. In order to counter the lack of data, it is important to contribute to the generation of new sets of images available to all researchers, to facilitate comparison between different algorithms. Therefore, one of the contributions of our paper is the introduction of a new and challenging dataset for semantic segmentation scenarios of fig plants, which is presented through
Section 3.
4. Convolutional Neural Network
In top-view images of a fig crop most of the leaves are overlapped and present different tonality due to the sunlight and shadows. Also, the leaves can be camouflaged with the weed. Thus, with an approach based on hand-engineered features, the expected result could hardly be obtained. On the contrary, it has been proven that a CNN has the capability to discover effective representations of complex scenes in order to perform good discrimination in different Computer Vision tasks with large image repositories. For these reasons, we decided to explore the CNN models to classify the pixels into crop or non-crop classes in order to perform a crop segmentation. In this section, we describe the CNN architecture for the segmentation of fig plants.
4.1. Approach
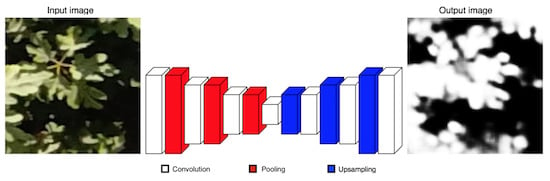
Our CNN is inspired by SegNet architecture [
14], which uses the principles of an encoder-decoder architecture to perform pixel-wise semantic segmentation. Unlike SegNet, our architecture comprises only 7 learnable layers as follows. The encoder section has 4 convolutional layers and 3 pooling layers to generate a low-resolution representation. The decoder part has 3 convolutional layers and 3 upsampling layers for producing pixel-wise predictions. In
Figure 3 we present a scheme of our approach. We use fewer convolutional layers and have a smaller number of trainable parameters than SegNet-Basic, the smaller version of SegNet, turning it into a simpler model. Likewise, we discard the multi-class soft-max classifier as a final layer because we expect only 2 classes. On the contrary, a sigmoid layer is used in the output to predict a probability of that each pixel belongs to one class or another.
The input of our CNN is a 128 × 128 RGB patch and the output is a 128 × 128 greyscale patch; details about image sizes are presented in the next section. For training, firstly each pixel value is normalized to a range of 0 to 1. The patch is passed through a set of different convolutional layers, where we use relatively large receptive fields (7 × 7) for the first layer and very small receptive fields (3 × 3) for the rest. The convolution stride is fixed to 1 pixel and a zero-padding option is used for all layers. Two activation functions are used. A Sigmoid activation is applied after the last convolution layer and for the rest, a Rectified Linear Unit (ReLU) activation is employed in order to introduce nonlinearities. Maxpooling is done over 2 × 2 windows with stride 2. Upsampling is performed by a factor of 2 × 2 to increase the resolution of the image. In
Table 2 we present a summary of our proposed CNN architecture. The convolutional layers parameters are denoted as “[receptive field size] |{
layer output} = [number of channels] [image size].”
The best set of hyperparameters that define the structure of the network (e.g., number of layers, number and size of filters) was determined by experience and performing a series of experiments. Concretely, we choose the following parameters: epochs = 120, batch size = 32, loss function = Binary cross-entropy, optimizer = Adadelta and initial learning rate = 1.0.
Our CNN is developed in Python using Keras [
31] and TensorFlow [
32] libraries. The experiments are performed on a laptop with a Linux platform, a processor Intel
® Core™ i7-8750H CPU @ 2.20GHz x 12, 16 GB RAM and NVIDIA GPU GeForce GTX 1070. We convert the final decoder output to a binary image in order to compute the metrics for evaluation by way of a simplest thresholding method. Values below 0.5 are turned to zero and all values above that threshold to 1.
4.2. Input Data Preparation
In our proposed data set, there are only 10 labelled images of 2000 × 1500 pixels. However, a fig leaf in these images is represented, on average, by a region of 25 × 25 pixels. Therefore, they contain a large number of leaves samples subjected to different conditions, with which it is possible to carry out training of our CNN without the need to resort to data augmentation techniques. We perform in each labelled image a sampling of overlapping patches with a fixed stride. The image is divided into patches of 128 × 128 pixels with horizontal and vertical overlapping between regions of 70% (90 pixels), generating more input data. We observe that as the size of the patch increases, the performance improves. Nevertheless, larger patches involve more processing time and the problem of getting a small number of patches per image. Finally, we work with a total of 19,380 patches with their respective GT.
6. Conclusions
We have proposed a fig plant segmentation method based on deep learning and a challenging data set with its ground truth labelled by hand at the pixel level. The data set is of particular interest to smart farming and computer vision researchers. It consists of 110 high resolution aerial images captured by an UAV. Images show an open field fig crop, where there is a great variability in tones and shapes of the leaves due to the plant morphology and the different positions of the camera relative to the sun when the image was captured. In addition, the background is really complex because it can contain several elements which increase the difficulty of the plant detection process. The fig species is Ficus carica, whose bushes are tall and whose life cycle is long, so the use of aerial robots is more appropriate than terrestrial ones for their monitoring.
Our approach was based on a CNN model with an encoder-decoder architecture trained end-to-end. The experimental results showed that our model can be trained in just 45 min while maintaining its ability to accurately segment the fig plants. The CNN-based method is adequate to deal with the two-class segmentation problem, even in highly challenging scenarios such as the segmentation of fig plants introduced in this work. The encoder-decoder architecture is capable to learn the discriminative filters that help detect fig foliage in order to segment them from the background. On the other hand, the evaluation of vegetation indices showed that ExG, CIVE, COM2 and MExG have an acceptable performance in our data, although it is clearly surpassed by the proposed convolutional encoder-decoder architecture. These indices can be used as first stage where it is necessary to isolate the vegetation as the object of interest quickly in order to do tasks of a higher level such as recognition or classification of plants. Future work is aimed at optimizing the model to improve results and consider other cases of fig crops in different seasons. Likewise, we plan to carry out experiments in orthomosaic images generated from our fig images. Orthomosaics are of great importance in agriculture because they offer more information, which could be used to analyse the conditions of the field.