Synergy of ICESat-2 and Landsat for Mapping Forest Aboveground Biomass with Deep Learning
Abstract
:1. Introduction
2. Materials and Methods
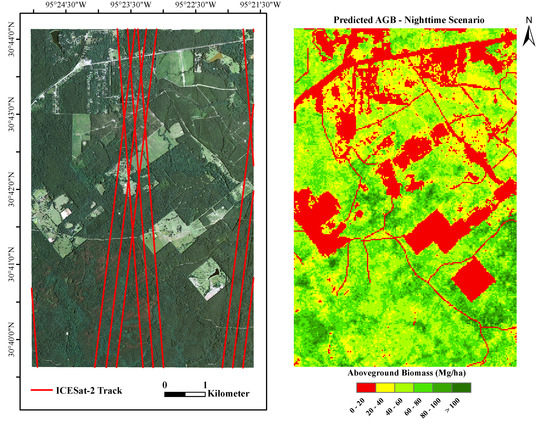
2.1. Study Area
2.2. Simulated ICESat-2-Estimated AGB
2.3. Mapped Predictors
- Spectral Metrics from Landsat 5 TM-
- ○
- Normalized Difference Vegetation Index (NDVI): (NIR − Red)/(NIR + Red)
- ○
- Enhanced Vegetation Index (EVI): 2.5 * ((NIR − Red)/(NIR + 6 * Red − 7.5 * Blue + 1))
- ○
- Soil Adjusted Vegetation Index (SAVI): ((NIR − Red)/(NIR + Red + 0.5)) * (1.5)
- ○
- Modified Soil Adjusted Vegetation Index (MSAVI): (2 * NIR + 1 − sqrt ((2 * NIR + 1)² − 8 * (NIR − Red)))/2
- NLCD 2011 land cover map
- NLCD 2011 US Forest Service tree canopy cover
2.4. Deep Neural Networks (DNNs)
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hall, F.G.; Bergen, K.; Blair, J.B.; Dubayah, R.; Houghton, R.; Hurtt, G.; Kellndorfer, J.; Lefsky, M.; Ranson, J.; Saatchi, S.; et al. Characterizing 3D vegetation structure from space: Mission requirements. Remote Sens. Environ. 2011, 115, 2753–2775. [Google Scholar] [CrossRef] [Green Version]
- Zald, H.S.J.; Wulder, M.A.; White, J.C.; Hilker, T.; Hermosilla, T.; Hobart, G.W.; Coops, N.C. Integrating Landsat pixel composites and change metrics with lidar plots to predictively map forest structure and aboveground biomass in Saskatchewan, Canada. Remote Sens. Environ. 2016, 176, 188–201. [Google Scholar] [CrossRef] [Green Version]
- Goetz, S.; Dubayah, R. Advances in remote sensing technology and implications for measuring and monitoring forest carbon stocks and change. Carbon Manag. 2011, 2, 231–244. [Google Scholar] [CrossRef]
- Chi, H.; Sun, G.Q.; Huang, J.L.; Guo, Z.F.; Ni, W.J.; Fu, A.M. National Forest Aboveground Biomass Mapping from ICESat/GLAS Data and MODIS Imagery in China. Remote Sens. 2015, 7, 5534–5564. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Su, Y.; Xue, B.; Liu, J.; Zhao, X.; Fang, J.; Guo, Q. Mapping Global Forest Aboveground Biomass with Spaceborne LiDAR, Optical Imagery, and Forest Inventory Data. Remote Sens. 2016, 8, 565. [Google Scholar] [CrossRef]
- Nelson, R.; Margolis, H.; Montesano, P.; Sun, G.; Cook, B.; Corp, L.; Andersen, H.-E.; deJong, B.; Pellat, F.P.; Fickel, T.; et al. Lidar-based estimates of aboveground biomass in the continental US and Mexico using ground, airborne, and satellite observations. Remote Sens. Environ. 2017, 188, 127–140. [Google Scholar] [CrossRef] [Green Version]
- Glenn, N.F.; Neuenschwander, A.; Vierling, L.A.; Spaete, L.; Li, A.H.; Shinneman, D.J.; Pilliod, D.S.; Arkle, R.S.; McIlroy, S.K. Landsat 8 and ICESat-2: Performance and potential synergies for quantifying dryland ecosystem vegetation cover and biomass. Remote Sens. Environ. 2016, 185, 233–242. [Google Scholar] [CrossRef]
- Popescu, S.C. Estimating biomass of individual pine trees using airborne lidar. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar] [CrossRef]
- Neuenschwander, A.; Pitts, K. The ATL08 land and vegetation product for the ICESat-2 Mission. Remote Sens. Environ. 2019, 221, 247–259. [Google Scholar] [CrossRef]
- Stysley, P.R.; Coyle, D.B.; Clarke, G.B.; Frese, E.; Blalock, G.; Morey, P.; Kay, R.B.; Poulios, D.; Hersh, M. Laser Production for NASA’s Global Ecosystem Dynamics Investigation (GEDI) Lidar. In Proceedings of the Conference on Laser Radar Technology and Applications XXI, Baltimore, MD, USA, 19–20 April 2016. [Google Scholar]
- Markus, T.; Neumann, T.; Martino, A.; Abdalati, W.; Brunt, K.; Csatho, B.; Farrell, S.; Fricker, H.; Gardner, A.; Harding, D.; et al. The Ice, Cloud, and land Elevation Satellite-2 (ICESat-2): Science requirements, concept, and implementation. Remote Sens. Environ. 2017, 190, 260–273. [Google Scholar] [CrossRef]
- Marselis, S.; Armston, J.; Dubayah, R. Summary of the Second GEDI Science Team Meeting. Earth Obs. 2016, 6, 31–36. [Google Scholar]
- Sun, X. Space-Based Lidar Systems. In Proceedings of the Conference on Lasers and Electro-Optics 2012, San Jose, CA, USA, 6–11 May 2012. [Google Scholar]
- Neuenschwander, A.L.; Magruder, L.A. The Potential Impact of Vertical Sampling Uncertainty on ICESat-2/ATLAS Terrain and Canopy Height Retrievals for Multiple Ecosystems. Remote Sens. 2016, 8, 16. [Google Scholar] [CrossRef]
- Shao, Z.F.; Zhang, L.J.; Wang, L. Stacked Sparse Autoencoder Modeling Using the Synergy of Airborne LiDAR and Satellite Optical and SAR Data to Map Forest Above-Ground Biomass. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 5569–5582. [Google Scholar] [CrossRef]
- Lu, D.S.; Chen, Q.; Wang, G.X.; Liu, L.J.; Li, G.Y.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Popescu, S.C.; Zhao, K.; Neuenschwander, A.; Lin, C. Satellite lidar vs. small footprint airborne lidar: Comparing the accuracy of aboveground biomass estimates and forest structure metrics at footprint level. Remote Sens. Environ. 2011, 115, 2786–2797. [Google Scholar] [CrossRef]
- Lefsky, M.A. A global forest canopy height map from the Moderate Resolution Imaging Spectroradiometer and the Geoscience Laser Altimeter System. Geophys. Res. Lett. 2010, 37, 5. [Google Scholar] [CrossRef]
- Simard, M.; Pinto, N.; Fisher, J.B.; Baccini, A. Mapping forest canopy height globally with spaceborne lidar. J. Geophys. Res. Biogeosci. 2011, 116, 12. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Baccini, A.; Friedl, M.A.; Woodcock, C.E.; Warbington, R. Forest biomass estimation over regional scales using multisource data. Geophys. Res. Lett. 2004, 31, 4. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, J.; Martinez-Alvarez, F.; Troncoso, A.; Riquelme, J.C. A comparison of machine learning regression techniques for LiDAR-derived estimation of forest variables. Neurocomputing 2015, 167, 24–31. [Google Scholar] [CrossRef]
- Tian, X.; Li, Z.; Su, Z.; Chen, E.; van der Tol, C.; Li, X.; Guo, Y.; Li, L.; Ling, F. Estimating montane forest above-ground biomass in the upper reaches of the Heihe River Basin using Landsat-TM data. Int. J. Remote Sens. 2014, 35, 7339–7362. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, J.P.; Xu, S.H.; Dong, J.J.; Yang, Y. Forest Above Ground Biomass Estimation from Remotely Sensed Imagery in the Mount Tai Area Using the RBF ANN Algorithm. Intell. Autom. Soft Comput. 2018, 24, 391–398. [Google Scholar] [CrossRef]
- Hatcher, W.G.; Yu, W. A Survey of Deep Learning: Platforms, Applications and Emerging Rlesearch Trends. IEEE Access 2018, 6, 24411–24432. [Google Scholar] [CrossRef]
- Lv, J.J.; Zhang, B.; Li, X.Q. Research on Object Detection Algorithm Based on PVANet. Adv. Comput. Commun. Comput. Sci. 2019, 759, 141–151. [Google Scholar] [CrossRef]
- Shafaey, M.A.; Salem, M.A.M.; Ebied, H.M.; Al-Berry, M.N.; Tolba, M.F. Deep Learning for Satellite Image Classification; Springer Nature: Basel, Switzerland, 2019. [Google Scholar]
- Zhu, X.X.; Tuia, D.; Mou, L.C.; Xia, G.S.; Zhang, L.P.; Xu, F.; Fraundorfer, F. Deep Learning in Remote Sensing. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, J.; Gonzalez-Ferreiro, E.; Mateos-Garcia, D.; Riquelme-Santos, J.C. A Preliminary Study of the Suitability of Deep Learning to Improve LiDAR-Derived Biomass Estimation. Hybrid. Artif. Intell. Syst. 2016, 9648, 588–596. [Google Scholar] [CrossRef]
- Trier, O.D.; Salberg, A.B.; Kermit, M.; Rudjord, O.; Gobakken, T.; Naesset, E.; Aarsten, D. Tree species classification in Norway from airborne hyperspectral and airborne laser scanning data. Eur. J. Remote Sens. 2018, 51, 336–351. [Google Scholar] [CrossRef]
- Neuenschwander, A.; Popescu, S.; Nelson, R.; Harding, D.; Pitts, K.; Pederson, D.; Sheridan, R. ICE, CLOUD, and Land Elevation Satellite (ICESat-2) Algorithm Theoretical Basis Document (ATBD) for Land —Vegetation Along-track products (ATL08). Available online: https://go.nasa.gov/31PqmKp (accessed on 24 June 2019).
- Baghdadi, N.; le Maire, G.; Fayad, I.; Bailly, J.S.; Nouvellon, Y.; Lemos, C.; Hakamada, R. Estimation of Forest Height and Aboveground Biomass from ICESat-2/GLAS Data in Eucaluptus Plantations in Brazil. In Proceedings of the 2014 IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 725–728. [Google Scholar] [CrossRef]
- Harding, D.J.; Carabajal, C.C. ICESat waveform measurements of within-footprint topographic relief and vegetation vertical structure. Geophys. Res. Lett. 2005, 32, 4. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Keller, M.; Pang, Y.; de Camargo, P.B.; Hunter, M.O. Revised method for forest canopy height estimation from Geoscience Laser Altimeter System waveforms. J. Appl. Remote Sens. 2007, 1, 18. [Google Scholar] [CrossRef]
- Liu, K.; Wang, J.; Zeng, W.; Song, J. Comparison and Evaluation of Three Methods for Estimating Forest above Ground Biomass Using TM and GLAS Data. Remote Sens. 2017, 9, 341. [Google Scholar] [CrossRef]
- Gwenzi, D.; Lefsky, M.A.; Suchdeo, V.P.; Harding, D.J. Prospects of the ICESat-2 laser altimetry mission for savanna ecosystem structural studies based on airborne simulation data. ISPRS J. Photogramm. Remote Sens. 2016, 118, 68–82. [Google Scholar] [CrossRef]
- Montesano, P.M.; Rosette, J.; Sun, G.; North, P.; Nelson, R.F.; Dubayah, R.O.; Ranson, K.J.; Kharuk, V. The uncertainty of biomass estimates from modeled ICESat-2 returns across a boreal forest gradient. Remote Sens. Environ. 2015, 158, 95–109. [Google Scholar] [CrossRef]
- Narine, L.L.; Popescu, S.; Neuenschwander, A.; Zhou, T.; Srinivasan, S.; Harbeck, K. Estimating aboveground biomass and forest canopy cover with simulated ICESat-2 data. Remote Sens. Environ. 2019, 224, 1–11. [Google Scholar] [CrossRef]
- Popescu, S.C.; Zhou, T.; Nelson, R.; Neuenschwander, A.; Sheridan, R.; Narine, L.; Walsh, K.M. Photon counting LiDAR: An adaptive ground and canopy height retrieval algorithm for ICESat-2 data. Remote Sens. Environ. 2018, 208, 154–170. [Google Scholar] [CrossRef]
- Narine, L.L.; Popescu, S.; Zhou, T.; Srinivasan, S.; Harbeck, K. Mapping forest aboveground biomass with a simulated ICESat-2 vegetation canopy product and Landsat data. Ann. For. Res. 2019, 62. [Google Scholar] [CrossRef]
- Stathakis, D. How many hidden layers and nodes? AU—Stathakis, D. Int. J. Remote Sens. 2009, 30, 2133–2147. [Google Scholar] [CrossRef]
- Homer, C.; Dewitz, J.; Yang, L.M.; Jin, S.; Danielson, P.; Xian, G.; Coulston, J.; Herold, N.; Wickham, J.; Megown, K. Completion of the 2011 National Land Cover Database for the Conterminous United States—Representing a Decade of Land Cover Change Information. Photogramm. Eng. Remote Sens. 2015, 81, 345–354. [Google Scholar] [CrossRef]
- Blair, J.B.; Hofton, M.A. Modeling laser altimeter return waveforms over complex vegetation using high-resolution elevation data. Geophys. Res. Lett. 1999, 26, 2509–2512. [Google Scholar] [CrossRef]
- Martino, A. ATLAS Performance Spreadsheet. Available online: http://icesat.gsfc.nasa.gov/icesat2/data/sigma/sigma_data.php (accessed on 4 June 2018).
- Moussavi, M.S.; Abdalati, W.; Scambos, T.; Neuenschwander, A. Applicability of an automatic surface detection approach to micropulse photon-counting lidar altimetry data: Implications for canopy height retrieval from future ICESat-2 data. Int. J. Remote Sens. 2014, 35, 5263–5279. [Google Scholar] [CrossRef]
- Snyder, K.A.; Huntington, J.L.; Wehan, B.L.; Morton, C.G.; Stringham, T.K. Comparison of Landsat and Land-Based Phenology Camera Normalized Difference Vegetation Index (NDVI) for Dominant Plant Communities in the Great Basin. Sensors 2019, 19, 1139. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning with Python; Manning Publications Co.: New York, NY, USA, 2018. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; p. 781. [Google Scholar]
- Bengio, Y. Learning Deep Architectures for AI; Now Publishers Inc.: Hanover, MA, USA, 2009; Volume 2, pp. 1–55. [Google Scholar]
- Doukim, C.A.; Dargham, J.A.; Chekima, A. Finding the number of hidden neurons for an MLP neural network using coarse to fine search technique. In Proceedings of the 10th International Conference on Information Science, Signal Processing and their Applications (ISSPA 2010), Kuala Lumpur, Malaysia, 10–13 May 2010; pp. 606–609. [Google Scholar]
- Guang-Bin, H. Learning capability and storage capacity of two-hidden-layer feedforward networks. IEEE Trans. Neural Netw. 2003, 14, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Wen, J.; Quitadamo, A.; Cheng, J.L.; Shi, X.H. A deep auto-encoder model for gene expression prediction. BMC Genom. 2017, 18, 39–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foody, G.M.; Cutler, M.E.; McMorrow, J.; Pelz, D.; Tangki, H.; Boyd, D.S.; Douglas, I. Mapping the biomass of Bornean tropical rain forest from remotely sensed data. Glob. Ecol. Biogeogr. 2001, 10, 379–387. [Google Scholar] [CrossRef]
- Li, L.; Guo, Q.H.; Tao, S.L.; Kelly, M.G.; Xu, G.C. Lidar with multi-temporal MODIS provide a means to upscale predictions of forest biomass. Isprs J. Photogramm. Remote Sens. 2015, 102, 198–208. [Google Scholar] [CrossRef]
- Ikasari, I.H.; Ayumi, V.; Fanany, M.I.; Mulyono, S. Multiple Regularizations Deep Learning for Paddy Growth Stages Classification from LANDSAT-8. In Proceedings of the 2016 International Conference on Advanced Computer Science and Information Systems, Malang, Indonesia, 15–16 October 2016; IEEE: New York, NY, USA, 2016; pp. 512–517. [Google Scholar]
- Rosen, P.; Hensley, S.; Shaffer, S.; Edelstein, W.; Kim, Y.; Kumar, R.; Misra, T.; Bhan, R.; Satish, R.; Sagi, R.; et al. An update on the NASA-ISRO dual-frequency DBF SAR (NISAR) Mission. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 2106–2108. [Google Scholar] [CrossRef]
- Carreiras, J.M.B.; Shaun, Q.G.; Toan, T.L.; Minh, D.H.T.; Saatchi, S.S.; Carvalhais, N.; Reichstein, M.; Scipal, K. Coverage of high biomass forests by the ESA BIOMASS mission under defense restrictions. Remote Sens. Environ. 2017, 196, 154–162. [Google Scholar] [CrossRef]
- Nunez, R.A.C.; de la Pena, M.; Irigollen, A.F.; Rodriguez, M.R. Deep learning models for the prediction of small-scale fisheries catches: Finfish fishery in the region of “Bahia Magadalena-Almejas”. ICES J. Mar. Sci. 2018, 75, 2088–2096. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D. Improved Prediction of Harmful Algal Blooms in Four Major South Korea’s Rivers Using Deep Learning Models. Int. J. Environ. Res. Public Health 2018, 15, 15. [Google Scholar] [CrossRef] [PubMed]








| Number of Neurons in 1st Hidden Layer | Daytime Scenario | Nighttime Scenario | No Noise Scenario | |||
|---|---|---|---|---|---|---|
| R2 | RMSE (Mg/ha) | R2 | RMSE (Mg/ha) | R2 | RMSE (Mg/ha) | |
| 20 | 0.40 | 19.95 | 0.45 | 19.97 | 0.46 | 20.60 |
| 40 | 0.40 | 20.01 | 0.45 | 19.98 | 0.46 | 20.55 |
| 60 | 0.40 | 19.98 | 0.46 | 19.92 | 0.47 | 20.49 |
| 80 | 0.40 | 19.94 | 0.46 | 19.91 | 0.47 | 20.49 |
| 100 | 0.40 | 19.94 | 0.45 | 19.99 | 0.47 | 20.44 |
| 120 | 0.40 | 19.95 | 0.45 | 19.94 | 0.47 | 20.46 |
| 140 | 0.40 | 20.00 | 0.46 | 19.89 | 0.47 | 20.44 |
| 160 | 0.40 | 20.02 | 0.45 | 19.97 | 0.47 | 20.38 |
| 180 | 0.39 | 20.08 | 0.46 | 19.84 | 0.47 | 20.43 |
| 200 | 0.39 | 20.06 | 0.45 | 19.98 | 0.47 | 20.46 |
| 300 | 0.40 | 19.90 | 0.46 | 19.88 | 0.47 | 20.36 |
| 400 | 0.39 | 20.09 | 0.46 | 19.85 | 0.47 | 20.32 |
| 500 | 0.39 | 20.05 | 0.45 | 20.08 | 0.48 | 20.29 |
| 600 | 0.40 | 19.91 | 0.47 | 19.72 | 0.47 | 20.34 |
| 700 | 0.39 | 20.13 | 0.46 | 19.87 | 0.47 | 20.37 |
| 800 | 0.40 | 19.93 | 0.46 | 19.75 | 0.47 | 20.35 |
| 900 | 0.40 | 19.98 | 0.46 | 19.84 | 0.47 | 20.36 |
| 1000 | 0.38 | 20.27 | 0.45 | 20.00 | 0.48 | 20.30 |
| Daytime Scenario Model Structure: 6-300-160-1 | Nighttime Scenario Model Structure: 6-600-400-1 | No Noise Scenario Model Structure: 6-500-300-60-1 | ||||
|---|---|---|---|---|---|---|
| Learning Rate | R2 | RMSE | R2 | RMSE | R2 | RMSE |
| 0.1 | 0.25 | 22.32 | 0.34 | 22.01 | 0.40 | 21.66 |
| 0.01 | 0.39 | 20.17 | 0.45 | 20.02 | 0.41 | 21.48 |
| 0.001 | 0.42 | 19.57 | 0.48 | 19.42 | 0.50 | 19.82 |
| 0.0001 | 0.42 | 19.55 | 0.49 | 19.35 | 0.50 | 19.82 |
| RF Model—6 Predictor Variables | RF Model—94 Predictor Variables | DNN Model—6 Predictor Variables | DNN Model—94 Predictor Variables | |||||
|---|---|---|---|---|---|---|---|---|
| Scenario | R² | RMSE | R² | RMSE | R² | RMSE | R² | RMSE |
| Daytime | 0.42 | 19.69 Mg/ha | 0.64 | 15.58 Mg/ha | 0.42 | 19.55 Mg/ha | 0.64 | 15.47 Mg/ha |
| Nighttime | 0.49 | 19.30 Mg/ha | 0.66 | 15.89 Mg/ha | 0.49 | 19.35 Mg/ha | 0.66 | 15.64 Mg/ha |
| No Noise | 0.51 | 19.72 Mg/ha | 0.68 | 15.93 Mg/ha | 0.50 | 19.82 Mg/ha | 0.67 | 16.09 Mg/ha |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narine, L.L.; Popescu, S.C.; Malambo, L. Synergy of ICESat-2 and Landsat for Mapping Forest Aboveground Biomass with Deep Learning. Remote Sens. 2019, 11, 1503. https://doi.org/10.3390/rs11121503
Narine LL, Popescu SC, Malambo L. Synergy of ICESat-2 and Landsat for Mapping Forest Aboveground Biomass with Deep Learning. Remote Sensing. 2019; 11(12):1503. https://doi.org/10.3390/rs11121503
Chicago/Turabian StyleNarine, Lana L., Sorin C. Popescu, and Lonesome Malambo. 2019. "Synergy of ICESat-2 and Landsat for Mapping Forest Aboveground Biomass with Deep Learning" Remote Sensing 11, no. 12: 1503. https://doi.org/10.3390/rs11121503
APA StyleNarine, L. L., Popescu, S. C., & Malambo, L. (2019). Synergy of ICESat-2 and Landsat for Mapping Forest Aboveground Biomass with Deep Learning. Remote Sensing, 11(12), 1503. https://doi.org/10.3390/rs11121503