1. Introduction
For many years, remote sensing (RS) systems have been widely applied for agricultural monitoring and crop identification [
1,
2,
3,
4], and these systems provide many surface reflectance images that can be utilized to derive hidden patterns of vegetation coverage. In crop identification tasks, the information of growing dynamics or sequential relationships derived from time-series images is used to perform classification. Although high-spatial-resolution datasets such as Landsat have clear advantages for capturing the fine spatial details of the land surface, such datasets typically do not have high temporal coverage frequency over large regions and are often badly affected by extensive cloud cover. However, coarse-resolution sensors such as the Moderate Resolution Imaging Spectroradiometer (MODIS) provide data at a near-daily observational coverage frequency and over large areas [
5]. While MODIS data are not a proper option for resolving smaller field sizes, they do provide a valuable balance between high temporal frequency and high spatial resolution [
6].
Many methods for crop identification or vegetation classification using MODIS time series have been examined and implemented in the academic world [
5,
6,
7]. A common approach for treating multitemporal data is to retrieve temporal features or phonological metrics from vegetation index series obtained by band calculations. According to phenology and simple statistics, several key phenology metrics, such as the base level, maximum level, amplitude, start date of the season, end date of the season, and length of the season, extracted from time-series RS images are used as classification features that are sufficient for accurate crop identification. For example, Cornelius Senf et al. [
5] mapped rubber plantations and natural forests in Xishuangbanna (Southwest China) using multispectral phenological metrics from MODIS time series, which achieved an overall accuracy of 0.735. They showed that the key phenological metrics discriminating rubber plantations and natural forests were the timing of seasonal events in the shortwaved infrared (SWIR) reflectance time series and the Enhanced Vegetation Index (EVI) or SWIR reflectance during the dry season. Pittman et al. [
6] estimated the global cropland extent and used the normalized difference vegetation index (NDVI) and thermal data to depict cropland phenology over the study period. Subpixel training datasets were used to generate a set of global classification tree models using a bagging methodology, resulting in a global per-pixel cropland probability layer. Tuanmu et al. [
7] used phenology metrics generated from MODIS time series to characterize the phenological features of forests with understory bamboo. Using maximum entropy modeling together with these phenology metrics, they successfully mapped the spatial distribution of understory bamboo. To address image noise such as pseudo-lows and pseudo-hikes caused by shadows, clouds, weather or sensors, many studies have used mathematical functions or complex models to smooth the vegetation index time series before feature extraction. Toshihiro Sakamoto et al. [
8] adopted wavelet and Fourier transforms for filtering time-series EVI data. Zhang et al. [
9] used a series of piecewise logistic functions fit to remotely sensed vegetation index data to represent intra-annual vegetation dynamics. Furthermore, weighted linear regression [
10], asymmetric Gaussian smoothing [
11,
12], Whittaker smoothing [
13], and Savitzky-Golay filtering [
14] have also been widely used for the same reason. Yang Shao et al. [
15] compared the Savitzky-Golay, asymmetric Gaussian, double-logistic, Whittaker, and discrete Fourier transformation smoothing algorithms (noise reduction) and applied them to MODIS NDVI time-series data to provide continuous phenology data for land cover classifications across the Laurentian Great Lakes Basin, proving that the application of a smoothing algorithm significantly reduced image noise compared to the raw data.
Although temporal feature extraction-based approaches have exhibited good performance in crop identification tasks, they have some weaknesses. First, general temporal features or phenological characteristics may not be appropriate for the specific task. Expert experience and domain knowledge are highly needed to design proper features and a feature extraction pipeline. Second, the features extracted from the time series cannot always fully utilize all the data, and information loss is inevitable. These types of feature extraction processes usually come with limitations in terms of automation and flexibility when considering large-scale classification tasks [
16].
Intelligent algorithms such as random forest (RF) and deep neural networks (DNNs) can learn directly from the original values of MODIS data. They can apply all the values of a time series as input and do not need well-designed feature extractors, which could prevent the information loss that often occurs in temporal feature extraction. These algorithms are convenient for large-scale implementation and application, and there are many processing frameworks based on RF or DNNs that are being implemented for cropland identification. Related works will be introduced in the next paragraphs.
Recently, RF classifiers have been widely used for RS images due to their explicit and explainable decision-making process, and these classifiers are easily implemented in a parallel structure for computing acceleration. Rodriguez-Galiano et al. [
17] explored the performance of an RF classifier in classifying 14 different land categories in the south of Spain. Results show that the RF algorithm yields accurate land cover classifications and is robust to the reduction of the training set size and noise compared to the classification trees. Charlotte Pelletier et al. [
18] assessed the robustness of using RF to map land cover and compared the algorithm with a support vector machine (SVM) algorithm. RF achieved an overall accuracy of 0.833, while SVM achieved an accuracy of 0.771. Works based on RF usually use a set of decision trees trained by different subsets of samples to make predictions collaboratively [
19]. However, the splitting nodes still used well-designed features with random selection, and this procedure might be complex and inefficient for the classification of large-scale areas. In this paper, we proposed an RF-based model that directly learns from the original values of the MODIS time series for large-scale crop identification in the Huanghuaihai Region to address the task in an efficient manner.
Considering the successful applications of deep learning (DL) in computer vision, deep models have also been evaluated for time-series image classification [
16,
20,
21]. Researchers usually use pretrained baseline architectures of convolutional neural networks (CNNs), such as AlexNet [
22], GoogLeNet [
23] and ResNet [
24], and fine-tuning to automatically obtain advanced representations of data, which are usually followed by a softmax layer or SVM to adapt to specific RS classification tasks. For times series scenarios, recurrent neural networks (RNNs) and long short-term memory (LSTM) networks are often used to analyze RS images due to their ability to capture long-term dependencies. Marc Rußwurm et al. [
20] employed LSTM networks to extract temporal characteristics from a sequence of SENTINEL 2A observations and compared the performance with SVM baseline architectures. Zhong et al. [
16] designed two types of DNN models for multitemporal crop classification: one was based on LSTM networks, and the other was based on one-dimensional convolutional (Conv1D) layers. Three widely used classifiers were also tested and compared, including gradient boosting machine (XGBoost), RF, and SVM classifiers. Although LSTM is widely used for sequential data representation, Zhong et al. [
16] revealed that its accuracy was the lowest among all the classifiers. Considering that the identification of crop types is highly dependent on a few temporal images of key growth stages such as the green-returning and jointing stages of winter wheat, it is important for the model to have the ability to pay attention to the critical images of the times series. In early studies, attention-based LSTM models were used to address sequence-to-sequence language translation tasks [
25], which could generate a proper word each time according to the specific input word and the context. Inspired by the machine translation community and crop growth cycle intuition, we proposed an attention-based LSTM model (A-LSTM) to identify winter wheat areas. The LSTM part of the model transforms original values to advanced representations and then follows an attention layer that encodes the sequence to one fixed-length vector that is used to decode the output at each timestep. A final softmax layer is then used to make a prediction.
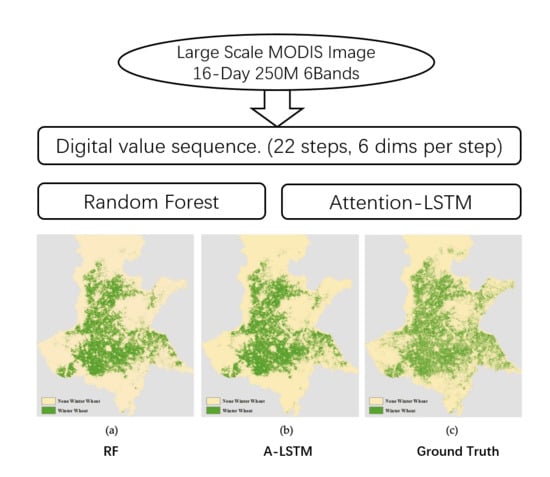
In this study, we proposed two models, RF and A-LSTM, that can be efficiently used for large-scale winter wheat identification throughout the Huanghuaihai Region, by building an automatic data preprocessing pipeline that transforms time-series MODIS tiles into training samples that can be directly fed into the models. As this study is the first to apply an attention mechanism-based LSTM model to the classification of time-series images, a comparison with RF and an evaluation of the performance were also conducted. In addition, we analyzed the impacts of the pixel-mixing effect with two different training strategies in this paper. Furthermore, with the intuition that there is some difference in wheat sowing and harvesting time from north to south, we also evaluated the generalizability of the models to different areas. Finally, our models were used to identify the distribution of winter wheat over the past year, and we evaluated the performance via visual interpretation. Finally, we discussed the advantages and disadvantages of our models.
5. Discussion
Our two models achieved F1 scores of 0.72 and 0.71 for identifying the winter wheat distribution in the Huanghuaihai Region. Previous works such as Senf et al. [
5], Tuanmu et al. [
7], Rodriguez-Galiano et al. [
17], Charlotte Pelletier et al. [
18], and Marc Rußwurm et al. [
20] regarding land cover classification or crop identification usually focused on only a small area, such as a county or city, which resulted in a classification map that was more detailed than ours. However, our trained models could also easily recognize the approximate winter wheat distribution, but in a large-scale area, and these models could be especially used in some close areas or historical cropland area identification. All the procedures are very simple to implement, and the results can be rapidly obtained.
Although the RF and A-LSTM methods achieved comparable performance according to our experiments, the computation costs are different. As the score tables above show, the overall accuracy and F1 score of RF is slightly higher than that of A-LSTM, but the computation time required to train an RF model is much greater than that required to train an A-LSTM model. As there are many samples throughout the study area, RF needs to traverse almost all samples and find the optimal split orders and split points to build a decision tree each time. In addition, it required almost 2 h to train a complete RF model on our 24-core working station with parallel computation. For A-LSTM, a high-performance graphics card could be used to accelerate the computation, and an early stop strategy [
34] (automatically stop training when the model converges) could be employed in practical applications. In this manner, approximately 50 min are required to train an A-LSTM model with a Tesla V100 GPU card.
Furthermore, the overall accuracies of our two models were 0.87 and 0.85. However, the precision and recall were approximately 0.70, which is not sufficiently precise for the detailed mapping of winter wheat. This result is mainly due to three phenomena. (1) the cell size of MODIS data is 250 m, and a cell might contain multiple land cover types, thus making the reflectance spectra unstable and unpredictable. (2) Our ground truth map, which was provided by the Chinese Academy of Agricultural Sciences, was assumed to be totally accurate in each cell. Since we did not receive any instructions regarding how to complete the map or information on the data accuracy, we reassessed the data via manually collected field data, as described in
Section 2.2.2. The results indicated that the overall accuracy of the ground truth map was 0.95, the precision of winter wheat was 0.89, and the recall was 0.83, which might result in noise in the training sample labels. (3) Although many works have demonstrated the effectiveness of RF and DNNs, they still have limitations in learning the characteristics of such a large and complex area. Using a more complex RF or A-LSTM (a larger number of trees with RF or a deeper network with A-LSTM) could increase the inference ability. However, this usually causes severe overfitting problems, and experiments have shown that the validation score remains almost unchanged when the models reach saturation.
In this paper, we also visualized the feature importance of RF. Generally, the features derived from March or April had high importance scores. In early March, the first joint emerges above the soil line. This joint provides visual evidence that the plants have initiated their reproduction [
35]. Then, winter wheat enters a fast-growing period until maturation. Thus, features in this period tend to be significant. For the feature importance of different bands, vegetation indices such as NDVI or EVI represent the reflectance differences between vegetation and soil in the visible and NIR bands. Green vegetation usually exhibits a large spectral difference between the red and NIR bands, thus making the vegetation index more important than a single band. In our study, features were derived from the six bands provided by the MOD13Q1, Collection 6 product. Future work could add additional bands in the models, which might provide better results.
When the trained models are used to make predictions in other areas, close areas usually have reliable results. The main reason behind this result is that the winter wheat growing season varies by area. Specifically, in the Huanghuaihai Region, winter wheat crops at the same latitudes likely have similar growing seasons. In addition, the experiments above support our point of view. Practically, there must be other reasons that result in the poor performance, such as the terrain, elevation, climate, crop species or different land cover compositions of the negative samples. Thus, the MODIS data distribution in the prediction area varies compared to that in the training areas. Our future work will focus on revealing the detailed mechanisms underlying this difference. Fortunately, when we applied our model to historical MODIS data, the performance was stable, as described in
Section 4.6. However, exceptions sometimes exist that result in noise in the prediction, such as improved winter wheat species, climate changes, and land cover changes. Regrettably, we did not conduct additional experiments over more past years because of the extensive labor required to collect validation data.
6. Conclusions
In this paper, we developed two models for large-scale winter wheat identification in the Huanghuaihai Region. To the best of our knowledge, this study was the first to use raw digital numbers derived from time-series MODIS images to implement classification pipelines and make predictions over a large-scale land area. According to our experiments, we can draw the following general conclusions. (1) Both the RF and A-LSTM models were efficient for identifying winter wheat areas, with F1 scores of 0.72 and 0.71, respectively. (2) The comparison of the two models indicates that RF achieved a slightly better score than A-LSTM, while A-LSTM is much faster with GPU acceleration. (3) For time-series winter wheat identification, the data acquired in March or April are more important and contribute more than the data acquired at other times. Vegetation indices such as EVI and NDVI are more helpful than single reflectance bands. (4) Predicting in local or nearby areas is more likely to yield reliable results. (5) The models are also capable of efficiently identifying historical winter wheat areas.
Our models were applied to only winter wheat identification due to the limitations of the crop distribution data, but they could potentially solve multiclass problems. Further research regarding multiple crop types or other land cover types over large regions could be more meaningful and useful. Moreover, due to the scarcity and acquisition difficulties of high-spatiotemporal-resolution images, the cell size of each training sample was 250 m, which is too large to avoid the mixed-pixel problem. We believe that using time-series images with finer scales could help improve the accuracy and generalizability of our models. Obviously, it is easy to determine how predictions are determined in RF models, but this information is difficult to determine in the A-LSTM model. Future research will likely devote more attention to the inference mechanisms of DNNs.