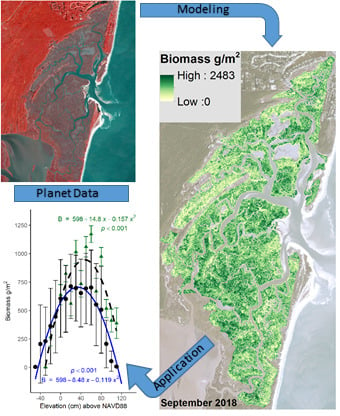
Estimating Aboveground Biomass and Its Spatial Distribution in Coastal Wetlands Utilizing Planet Multispectral Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Field Data Collection
2.3. Satellite Data
2.4. Vegetation Indicies
2.5. Statistical Analysis
3. Results
3.1. North Inlet
3.2. Plum Island
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Möller, I.; Kudella, M.; Rupprecht, F.; Spencer, T.; Paul, M.; van Wesenbeeck, B.K.; Wolters, G.; Jensen, K.; Bouma, T.J.; Miranda-Lange, M.; et al. Wave attenuation over coastal salt marshes under storm surge conditions. Nat. Geosci. 2014, 7, 727–731. [Google Scholar] [CrossRef] [Green Version]
- Narayan, S.; Beck, M.W.; Wilson, P.; Thomas, C.J.; Guerrero, A.; Shepard, C.C.; Reguero, B.G.; Franco, G.; Ingram, J.C.; Trespalacios, D. The Value of Coastal Wetlands for Flood Damage Reduction in the Northeastern USA. Sci. Rep. 2017, 7, 9463. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.; Acreman, M. Wetland nutrient removal: a review of the evidence. Hydrol. Earth Syst. Sci. 2004, 8, 673–685. [Google Scholar] [CrossRef] [Green Version]
- Nellemann, C.; Corcoran, E.; Duarte, C.M.; Valdés, L.; De Young, C.; Fonseca, L.; Grimsditch, G. Blue Carbon—The Role of Healthy Oceans in Binding Carbon; United Nations Environment Programme, GRID-Arendal: Nairobi, Kenya, 2009; ISBN 9788277010601. [Google Scholar]
- Mcleod, E.; Chmura, G.L.; Bouillon, S.; Salm, R.; Björk, M.; Duarte, C.M.; Lovelock, C.E.; Schlesinger, W.H.; Silliman, B.R. A blueprint for blue carbon: toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2. Front. Ecol. Environ. 2011, 9, 552–560. [Google Scholar] [CrossRef]
- Morris, J.T.; Sundareshwar, P.V.; Nietch, C.T.; Kjerfve, B. Responses of Coastal Wetlands to Rising Sea Level. Ecol. Soc. Am. 2002, 83, 2869–2877. [Google Scholar] [CrossRef]
- DeLaune, R.D.; Smith, C.J.; Patrick, W.H. Relationship of Marsh Elevation, Redox Potential, and Sulfide to Spartina alterniflora Productivity1. Soil Sci. Soc. Am. J. 1983, 47, 930. [Google Scholar] [CrossRef]
- Miller, G.J.; Morris, J.T.; Wang, C. Mapping salt marsh dieback and condition in South Carolina’s North Inlet-Winyah Bay National Estuarine Research Reserve using remote sensing. AIMS Environ. Sci. 2017, 4, 677–689. [Google Scholar] [CrossRef]
- Morris, J.T.; Sundberg, K.; Hopkinson, C.S. Salt Marsh Primary Production and Its Response to Relative Sea Level and Nutrients in Estuaries at Plum Island, Massachusets, and North Inlet, South Carolina, USA. Oceanography 2013, 26, 78–84. [Google Scholar] [CrossRef]
- Silliman, B.R.; Bertness, M.D. A trophic cascade regulates salt marsh primary production. Proc. Natl. Acad. Sci. USA 2002, 99, 10500–10505. [Google Scholar] [CrossRef] [Green Version]
- Mendelssohn, I.A. Nitrogen Metabolism in the Height Forms of Spartina Alterniflora in North Carolina. Ecology 1979, 60, 574–584. [Google Scholar] [CrossRef]
- Mendelssohn, I.A.; Seneca, E.D. The influence of soil drainage on the growth of salt marsh cordgrass Spartina alterniflora in North Carolina. Estuar. Coast. Mar. Sci. 1980. [Google Scholar] [CrossRef]
- Gross, M.F.; Klemas, V.; Hardisky, M.A. Long-term remote monitoring of salt marsh biomass. Remote Sens. Biosph. 1990, 1300, 59–70. [Google Scholar]
- Lumbierres, M.; Méndez, P.; Bustamante, J.; Soriguer, R.; Santamaría, L. Modeling Biomass Production in Seasonal Wetlands Using MODIS NDVI Land Surface Phenology. Remote Sens. 2017, 9, 392. [Google Scholar] [CrossRef]
- Gross, M.F.; Hardisky, M.A.; Klemas, V.; Wolf, P.L. Quantification of Biomass of the Marsh Grass Spartina altfernifara Loisel Using Landsat Thematic Mapper Imagery. Photogramm. Eng. Remote Sens. 1987, 53, 1577–1583. [Google Scholar]
- Mo, Y.; Kearney, M.S.; Riter, J.C.A. Post-deepwater horizon oil spill monitoring of Louisiana salt marshes using landsat imagery. Remote Sens. 2017, 9, 547. [Google Scholar] [CrossRef]
- Lopes, C.L.; Mendes, R.; Caçador, I.; Dias, J.M. Evaluation of long-term estuarine vegetation changes through Landsat imagery. Sci. Total Environ. 2019, 653, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Planet Team Planet Application Program Interface: In Space for Life on Earth. Available online: https://api.planet.com/ (accessed on 2 December 2018).
- Morris, J.T.; Haskin, B. A 5-yr Record of Aerial Primary Production and Stand Characteristics of Spartina Alterniflora. Ecology 1990, 71, 2209. [Google Scholar] [CrossRef]
- Morris, J.T. Effects of sea-level anomalies on estuarine processes. In Estuarine Science: A Synthetic Approach to Research and Practice; Hobbie, J.E., Ed.; Island Press: Washington, DC, USA, 2000; pp. 107–127. [Google Scholar]
- Pershing, A.J.; Alexander, M.A.; Hernandez, C.M.; Kerr, L.A.; Le Bris, A.; Mills, K.E.; Nye, J.A.; Record, N.R.; Scannell, H.A.; Scott, J.D.; et al. Slow adaptation in the face of rapid warming leads to collapse of the Gulf of Maine cod fishery. Science 2015, 350, 809–812. [Google Scholar] [CrossRef] [Green Version]
- NOAA Boston Tide Gage, Tides & Currents. Available online: https://tidesandcurrents.noaa.gov/stationhome.html?id=8443970 (accessed on 30 July 2019).
- Schwing, F.B.; Kjerfve, B.; Kjerfve, B. Longitudinal Characterization of a Tidal Marsh Creek Separating Two Hydrographically Distinct Estuaries. Estuaries 1980, 3, 236. [Google Scholar] [CrossRef]
- McKee, K.L.; Patrick, W.H. The Relationship of Smooth Cordgrass (Spartina alterniflora) to Tidal Datums: A Review. Estuaries 1988, 11, 143. [Google Scholar] [CrossRef]
- Cahoon, D.R.; Lynch, J.C.; Hensel, P.; Boumans, R.; Perez, B.C.; Segura, B.; Day, J.W. High-Precision Measurements of Wetland Sediment Elevation: I. Recent Improvements to the Sedimentation-Erosion Table. J. Sediment. Res. 2002, 72, 730–733. [Google Scholar] [CrossRef]
- Boumans, R.M.J.; Day, J.W. High Precision Measurements of Sediment Elevation in Shallow Coastal Areas Using a Sedimentation-Erosion Table. Estuaries 1993, 16, 375. [Google Scholar] [CrossRef]
- Davis, J.; Currin, C.; Morris, J.T. Impacts of Fertilization and Tidal Inundation on Elevation Change in Microtidal, Low Relief Salt Marshes. Estuaries and Coasts 2017, 40, 1677–1687. [Google Scholar] [CrossRef]
- Byrd, K.B.; O’Connell, J.L.; Di Tommaso, S.; Kelly, M. Evaluation of sensor types and environmental controls on mapping biomass of coastal marsh emergent vegetation. Remote Sens. Environ. 2014, 149, 166–180. [Google Scholar] [CrossRef]
- Li, W.; Gong, P. Continuous monitoring of coastline dynamics in western Florida with a 30-year time series of Landsat imagery. Remote Sens. Environ. 2016, 179, 196–209. [Google Scholar] [CrossRef]
- O’Donnell, J.; Schalles, J.; O’Donnell, J.P.R.; Schalles, J.F. Examination of Abiotic Drivers and Their Influence on Spartina alterniflora Biomass over a Twenty-Eight Year Period Using Landsat 5 TM Satellite Imagery of the Central Georgia Coast. Remote Sens. 2016, 8, 477. [Google Scholar] [CrossRef]
- Planet Team Planet Imagery Product Specification. Available online: https://assets.planet.com/docs/combined-imagery-product-spec-final-may-2019.pdf (accessed on 20 May 2019).
- Rouse, J.W.; Hass, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the great plains with ERTS. In Proceedings of the Third Earth Resources Technology Satellite (ERTS) symposium, Washington, DC, USA, 10–14 December; pp. 309–317.
- Hardisky, M.A.; Smart, R.M.; Klemas, V. Seasonal Spectral Characteristics and aboveground Biomass of the Tidal Marsh Plant, Spartina alterniflora. Photogramm. Eng. Remote Sens. 1983, 49, 85–92. [Google Scholar]
- Huete, A. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A Modified Soil Adjusted Vegetation Index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Roujean, J.-L.; Breon, F.-M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Wang, X.; Wang, M.; Wang, S.; Wu, Y. Extraction of vegetation information from visible unmanned aerial vehicle images. Trans. Chinese Soc. Agric. Eng. 2015, 31, 152–159. [Google Scholar]
- Gitelson, A.A.; Merzlyak, M.N. Remote sensing of chlorophyll concentration in higher plant leaves. Adv. Space Res. 1998, 22, 689–692. [Google Scholar] [CrossRef]
- R Core Team R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2002; ISBN 0387953647. [Google Scholar]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Willmott, C.J. Some Comments on the Evaluation of Model Performance. Bull. Am. Meteorol. Soc. 1982, 63, 1309–1313. [Google Scholar] [CrossRef] [Green Version]
- Zambrano-Bigiarini, M. hydroGOF: Goodness-of-fit functions for comparison of simulated and observed hydrological time series. Available online: http://hzambran.github.io/hydroGOF/ (accessed on 21 June 2019).
- Hardisky, M.A.; Daiber, F.C.; Roman, C.T.; Klemas, V. Remote sensing of biomass and annual net aerial primary productivity of a salt marsh. Remote Sens. Environ. 1984, 16, 91–106. [Google Scholar] [CrossRef]
- Yebra, M.; Chuvieco, E. Linking ecological information and radiative transfer models to estimate fuel moisture content in the Mediterranean region of Spain: Solving the ill-posed inverse problem. Remote Sens. Environ. 2009, 113, 2403–2411. [Google Scholar] [CrossRef]
- García, M.; Riaño, D.; Chuvieco, E.; Danson, F.M. Estimating biomass carbon stocks for a Mediterranean forest in central Spain using LiDAR height and intensity data. Remote Sens. Environ. 2010, 114, 816–830. [Google Scholar] [CrossRef]
- de Almeida, D.R.A.; Nelson, B.W.; Schietti, J.; Gorgens, E.B.; Resende, A.F.; Stark, S.C.; Valbuena, R. Contrasting fire damage and fire susceptibility between seasonally flooded forest and upland forest in the Central Amazon using portable profiling LiDAR. Remote Sens. Environ. 2016, 184, 153–160. [Google Scholar] [CrossRef]
- Valbuena, R.; Hernando, A.; Manzanera, J.A.; Görgens, E.B.; Almeida, D.R.A.; Silva, C.A.; García-Abril, A. Evaluating observed versus predicted forest biomass: R-squared, index of agreement or maximal information coefficient? Eur. J. Remote Sens. 2019, 52, 1–14. [Google Scholar] [CrossRef]
- Palmer, W.C. Meteorological drought. Res. Pap. 1965, 45, 58. [Google Scholar]
- NOAA National Climatic Data Center. Available online: https://www7.ncdc.noaa.gov/CDO/CDODivisionalSelect.jsp# (accessed on 26 May 2019).
- Dame, R.F.; Kenny, P.D. Variability of Spartina alterniflora primary production in the euhaline North Inlet estuary. Mar. Ecol. Prog. Ser. 1986, 32, 71–80. [Google Scholar] [CrossRef]
- Tyler, A.C.; Zieman, J.C. Patterns of development in the creekbank region of a barrier island Spartina alterniflora marsh. Mar. Ecol. Prog. Ser. 1999, 180, 161–177. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Pennings, S.C. Timing of disturbance affects biomass and flowering of a saltmarsh plant and attack by stem-boring herbivores. Ecosphere 2017, 8, e01675. [Google Scholar] [CrossRef]
- Walters, D.C.; Kirwan, M.L. Optimal hurricane overwash thickness for maximizing marsh resilience to sea level rise. Ecol. Evol. 2016, 6, 2948–2956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, Z.-T.; Gao, Y.; Xie, X.; Guo, H.-Q.; Zhang, T.-T.; Zhao, B. Spectral Discrimination of the Invasive Plant Spartina alterniflora at Multiple Phenological Stages in a Saltmarsh Wetland. PLoS ONE 2013, 8, e67315. [Google Scholar] [CrossRef] [PubMed]
- Jialin, L.; Hanbing, Z.; Xiaoping, Y. Study on the seasonal dynamics of zonal vegetation of NDVI/EVI of costal zonal vegetation based on MODIS data: A case study of Spartina alterniflora salt marsh on Jiangsu Coast, China. Afr. J. Agric. Res. 2011, 6, 4019–4024. [Google Scholar]









| Characteristic | Value |
|---|---|
| Blue wavelength (nm) | 455–415 |
| Green wavelength (nm) | 500–590 |
| Red wavelength (nm) | 590–670 |
| NIR wavelength (nm) | 780–860 |
| Spatial Resolution (m) | 3 × 3 |
| Temporal Resolution | Near daily |
| Image size (km) | 24 × 7 |
| Vegetation Index | Equation | Reference |
|---|---|---|
| NDVI | [32] | |
| SAVI 1 | [34] | |
| MSAVI2 | [35] | |
| RDVI | [36] | |
| VDVI | [37] | |
| GNDVI | [38] |
| Model | AIC | AICw |
|---|---|---|
| (MSAVI2) + (VDVI) | 364.4 | 0.3848 |
| MSAVI2 + VDVI | 365.2 | 0.2636 |
| SAVI + VDVI | 365.6 | 0.2122 |
| VDVI + GNDVI | 367.3 | 0.0856 |
| VDVI | 370.4 | 0.0231 |
| (B3) + (VDVI) | 371.8 | 0.0096 |
| B3 + (VDVI) | 372.3 | 0.0073 |
| MSAVI2 + B3 | 372.6 | 0.0064 |
| (MSAVI2) + (B3) | 373.3 | 0.0045 |
| SAVI + NDVI | 374.5 | 0.0025 |
| MSAVI2 | 382.4 | 0.0001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, G.J.; Morris, J.T.; Wang, C. Estimating Aboveground Biomass and Its Spatial Distribution in Coastal Wetlands Utilizing Planet Multispectral Imagery. Remote Sens. 2019, 11, 2020. https://doi.org/10.3390/rs11172020
Miller GJ, Morris JT, Wang C. Estimating Aboveground Biomass and Its Spatial Distribution in Coastal Wetlands Utilizing Planet Multispectral Imagery. Remote Sensing. 2019; 11(17):2020. https://doi.org/10.3390/rs11172020
Chicago/Turabian StyleMiller, Gwen J., James T. Morris, and Cuizhen Wang. 2019. "Estimating Aboveground Biomass and Its Spatial Distribution in Coastal Wetlands Utilizing Planet Multispectral Imagery" Remote Sensing 11, no. 17: 2020. https://doi.org/10.3390/rs11172020
APA StyleMiller, G. J., Morris, J. T., & Wang, C. (2019). Estimating Aboveground Biomass and Its Spatial Distribution in Coastal Wetlands Utilizing Planet Multispectral Imagery. Remote Sensing, 11(17), 2020. https://doi.org/10.3390/rs11172020