Improved Spatial-Spectral Superpixel Hyperspectral Unmixing
Abstract
:1. Introduction
2. The Linear Mixing Model (LMM)
3. Illustrating Spatial-Spectral Interaction
4. HSI Representation Using Superpixels
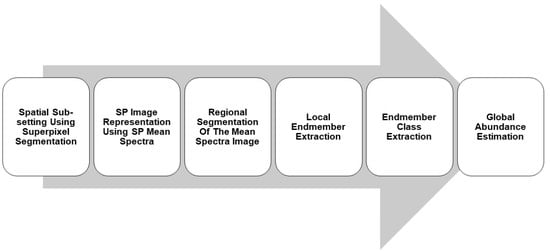
5. Proposed Unmixing Approach
5.1. Quadtree Regional Segmentation
5.2. Endmember Extraction
5.2.1. Estimating the Number of Endmembers
5.2.2. Endmember Extraction Using SVDSS
- Unfold the hyperspectral image cube (3D-array) into a matrix representation , where each column of X corresponds to the spectral signature of each image pixel, and N is the number of pixels.
- Compute the first p right singular vectors of X, .
- Let XP = [], where and . will be the matrix of endmembers
5.3. Endmember Class Extraction
5.4. Abundance Estimation
6. Experimental Results
6.1. Data Sets
6.1.1. HYDICE Urban Data Set
6.1.2. ROSIS Pavia University
6.2. Assessment Approach
Comparing Generated and Reference Maps
6.3. Experimental Results for the Urban Data Set
6.3.1. Qualitative Assessment
6.3.2. Quantitative Assessment
6.4. Experimental Results for the Pavia University Data Set
6.4.1. Qualitative Assessment
6.4.2. Quantitative Assessment
7. Comparing with Other Methods
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Keshava, N.; Mustard, J.F. Spectral unmixing. IEEE Signal Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Parente, M.; Plaza, A. Survey of geometric and statistical unmixing algorithms for hyperspectral images. In Proceedings of the 2nd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Reykjavik, Iceland, 14–16 June 2010; pp. 1–4. [Google Scholar]
- Heylen, R.; Parente, M.; Gader, P. A review of nonlinear hyperspectral unmixing methods. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1844–1868. [Google Scholar] [CrossRef]
- Keshava, N. A survey of spectral unmixing algorithms. Linc. Lab. J. 2003, 14, 55–78. [Google Scholar]
- Boardman, J.W. Automating spectral unmixing of AVIRIS data using convex geometry concepts. In Proceedings of the Summaries of the 4th Annual JPL Airborne Geoscience Workshop, Washington, DC, USA, 25–29 October 1993; Volume 1, pp. 11–14. [Google Scholar]
- Schott, J.R. Remote Sensing: The Image Chain Approach; Oxford University Press on Demand: Oxford, UK, 2007. [Google Scholar]
- Winter, M.E. N-FINDR: An algorithm for fast autonomous spectral end-member determination in hyperspectral data. Proc. SPIE 1999, 3753, 266–276. [Google Scholar]
- Nascimento, J.M.; Dias, J.M. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Schott, J.R.; Lee, K.; Raqueno, R.; Hoffmann, G.; Healey, G. A subpixel target detection technique based on the invariance approach. In Proceedings of the 12th JPL Airborne Earth Science Workshop, Pasadena, CA, USA, 25–28 February 2003; pp. 241–250. [Google Scholar]
- Plaza, A.; Martínez, P.; Pérez, R.; Plaza, J. Spatial/spectral endmember extraction by multidimensional morphological operations. IEEE Trans. Geosci. Remote Sens. 2002, 40, 2025–2041. [Google Scholar] [CrossRef] [Green Version]
- Torres-Madronero, M.C.; Velez-Reyes, M. Integrating spatial information in unsupervised unmixing of hyperspectral imagery using multiscale representation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1985–1993. [Google Scholar] [CrossRef]
- Shi, C.; Wang, L. Incorporating spatial information in spectral unmixing: A review. Remote Sens. Environ. 2014, 149, 70–87. [Google Scholar] [CrossRef]
- Rogge, D.M.; Rivard, B.; Zhang, J.; Sanchez, A.; Harris, J.; Feng, J. Integration of spatial–spectral information for the improved extraction of endmembers. Remote Sens. Environ. 2007, 110, 287–303. [Google Scholar] [CrossRef]
- Canham, K.; Schlamm, A.; Ziemann, A.; Basener, B.; Messinger, D. Spatially adaptive hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4248–4262. [Google Scholar] [CrossRef]
- Messinger, D.W.; Ziemann, A.K.; Schlamm, A.; Basener, W. Metrics of spectral image complexity with application to large area search. Opt. Eng. 2012, 51, 036201. [Google Scholar] [CrossRef]
- Messinger, D.; Ziemann, A.; Schlamm, A.; Basener, B. Spectral image complexity estimated through local convex hull volume. In Proceedings of the 2nd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Reykjavik, Iceland, 14–16 June 2010; pp. 1–4. [Google Scholar]
- Goenaga, M.A.; Torres-Madronero, M.C.; Velez-Reyes, M.; Van Bloem, S.J.; Chinea, J.D. Unmixing analysis of a time series of Hyperion images over the Guánica dry forest in Puerto Rico. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 6, 329–338. [Google Scholar] [CrossRef]
- Goenaga-Jimenez, M.A.; Velez-Reyes, M. Incorporating local information in unsupervised hyperspectral unmixing. Proc. SPIE 2012, 8390, 83901N. [Google Scholar]
- Goenaga-Jimenez, M.A.; Velez-Reyes, M. Integrating spatial information in unmixing using the nonnegative matrix factorization. Proc. SPIE 2014, 9088, 908811. [Google Scholar]
- Goenaga-Jimenez, M.A.; Velez-Reyes, M. Comparing quadtree region partitioning metrics for hyperspectral unmixing. Proc. SPIE 2013, 8743, 87430Z. [Google Scholar]
- Masalmah, Y.M.; Velez-Reyes, M. A full algorithm to compute the constrained positive matrix factorization and its application in unsupervised unmixing of hyperspectral imagery. Proc. SPIE 2008, 6966, 69661C. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef]
- Velez-Reyes, M.; Rosario, S. Solving adundance estimation in hyperspectral unmixing as a least distance problem. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Anchorage, AK, USA, 20–24 September 2004; Volume 5, pp. 3276–3278. [Google Scholar]
- Schowengerdt, R.A. Remote Sensing: Models and Methods for Image Processing; Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Zare, A.; Gader, P. PCE: Piecewise convex endmember detection. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2620–2632. [Google Scholar] [CrossRef]
- Cipar, J.J.; Lockwood, R.; Cooley, T.; Grigsby, P. Background spectral library for Fort A.P. Hill, Virginia. Proc. SPIE 2004, 5544, 35–47. [Google Scholar]
- Shlens, J. A tutorial on principal component analysis. arXiv 2014, arXiv:1404.1100. [Google Scholar]
- Zhang, X.; Chew, S.E.; Xu, Z.; Cahill, N.D. SLIC superpixels for efficient graph-based dimensionality reduction of hyperspectral imagery. Proc. SPIE 2015, 9472, 947209. [Google Scholar]
- Lézoray, O.; Meurie, C.; Celebi, M.E. Special Section Guest Editorial: Superpixels for Image Processing and Computer Vision. J. Electron. Imaging 2017, 26, 1. [Google Scholar] [CrossRef]
- Vedaldi, A.; Fulkerson, B. Vlfeat: An Open and Portable Library of Computer Vision Algorithms. In Proceedings of the 18th ACM International Conference on Multimedia, Firenze, Italy, 25–29 October 2010; 1469–1472; ACM: New York, NY, USA, 2010. [Google Scholar] [CrossRef]
- Thompson, D.R.; Mandrake, L.; Gilmore, M.S.; Castano, R. Superpixel endmember detection. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4023–4033. [Google Scholar] [CrossRef]
- Saranathan, A.M.; Parente, M. Uniformity-based superpixel segmentation of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1419–1430. [Google Scholar] [CrossRef]
- Yi, J.; Velez-Reyes, M. Dimensionality reduction using superpixel segmentation for hyperspectral unmixing using the cNMF. Proc. SPIE 2017, 10198, 101981H. [Google Scholar]
- Roger, A.; Horn, C.R.J. Matrix Analysis; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Suetin, P.; Kostrikin, A.I.; Manin, Y.I. Linear Algebra and Geometry; CRC Press: Boca Raton, FL, USA, 1997; Volume 1. [Google Scholar]
- Golub, G.H. Matrix Computations; Johns Hopkins University Press: Baltimore, MD, USA, 2013. [Google Scholar]
- Velez-Reyes, M.; Jimenez, L.O. Subset selection analysis for the reduction of hyperspectral imagery. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium Proceedings (IGARSS), Seattle, WA, USA, 6–10 July 1998; Volume 3, pp. 1577–1581. [Google Scholar]
- Aldeghlawi, M.; Velez-Reyes, M. Column Subset Selection Methods for Endmember Extraction in Hyperspectral Unmixing. Proc. SPIE 2018, 10644. [Google Scholar] [CrossRef]
- Mathworks. Statistics and Machine Learning Toolbox™ User’s Guide. 2017. Available online: https://www.mathworks.com/help/pdfdoc/stats/stats.pdf (accessed on 12 October 2019).
- Rosario-Torres, S.; Velez-Reyes, M. An algorithm for fully constrained abundance estimation in hyperspectral unmixing. Proc. SPIE 2005, 5806, 711–720. [Google Scholar]
- Zhu, F. Hyperspectral Unmixing: Ground Truth Labeling, Datasets, Benchmark Performances and Survey. Technical Report. arXiv 2017, arXiv:1708.05125. [Google Scholar]
- Zhu, F.; Wang, Y.; Fan, B.; Meng, G.; Pan, C. Effective Spectral Unmixing via Robust Representation and Learning-based Sparsity. arXiv 2014, arXiv:1409.0685. [Google Scholar]
- Plaza, A.; Martínez, P.; Perez, R.; Plaza, J. A comparative analysis of endmember extraction algorithms using AVIRIS hyperspectral imagery. In Proceedings of the Summaries of the 11th JPL Airborne Earth Science Workshop, Pasadena, CA, USA, 5–8 March 2001; pp. 267–276. [Google Scholar]
- Kuhnert, M.; Voinov, A.; Seppelt, R. Comparing raster map comparison algorithms for spatial modeling and analysis. Photogramm. Eng. Remote Sens. 2005, 71, 975–984. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Vedaldi, A.; Fulkerson, B. VLFeat: An Open and Portable Library of Computer Vision Algorithms. Available online: http://www.vlfeat.org/ (accessed on October 12 2019).
































| Reference Data | ||||||||
|---|---|---|---|---|---|---|---|---|
| Road | Grass | Trees | Roof | Dirt | Totals | User’s Agreement (%) | ||
| Class Map | Road | 13,775 | 296 | 368 | 96 | 238 | 14,773 | 93.24 |
| Grass | 44 | 29,443 | 1162 | 34 | 223 | 30,906 | 95.27 | |
| Trees | 59 | 2633 | 20,068 | 188 | 31 | 22,979 | 87.33 | |
| Roof | 1569 | 400 | 2108 | 6106 | 871 | 11,054 | 55.24 | |
| Dirt | 717 | 1542 | 736 | 281 | 5543 | 8819 | 62.85 | |
| Un-Assigned | 1301 | 2197 | 1647 | 222 | 347 | 5714 | ||
| Totals | 17,465 | 36,511 | 26,089 | 6927 | 7253 | 94,245 | ||
| Producer’s agreement (%) | 78.87 | 80.64 | 76.92 | 88.15 | 76.42 | Overall agreement = 79.51% | ||
| Harmonic Mean (%) | 85.46 | 87.35 | 81.80 | 67.92 | 68.98 | Kappa Statistic = 73.06 % | ||
| Reference Data | ||||||||
|---|---|---|---|---|---|---|---|---|
| Road | Grass | Trees | Roof | Dirt | Totals | User’s Agreement (%) | ||
| Class Map | Road | 15,369 | 1187 | 758 | 88 | 345 | 17747 | 86.6 |
| Grass | 248 | 32,484 | 458 | 20 | 156 | 33,366 | 97.36 | |
| Trees | 442 | 1968 | 23,678 | 362 | 145 | 26,595 | 89.03 | |
| Roof | 1055 | 257 | 1082 | 6387 | 1086 | 9867 | 64.73 | |
| Dirt | 169 | 365 | 85 | 69 | 5510 | 6198 | 88.90 | |
| Un-Assigned | 182 | 250 | 28 | 1 | 11 | 472 | ||
| Totals | 17,465 | 36,511 | 26,089 | 6927 | 7253 | 94,245 | ||
| Producer’s agreement (%) | 88.00 | 88.97 | 90.76 | 92.20 | 75.97 | Overall agreement = 88.52% | ||
| Harmonic Mean (%) | 87.29 | 92.97 | 89.89 | 76.06 | 81.93 | Kappa Statistic = 84.43 % | ||
| Reference Data | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Asphalt and Bitumen | Meadows and Soil | Gravel and Brick | Trees | Metal Sheet | Shadow | Totals | User’s Agreement (%) | ||
| Class Map | Asphalt and Bitumen | 3933 | 190 | 497 | 2 | 6 | 112 | 4740 | 82.97 |
| Meadows and Soil | 28 | 19,012 | 16 | 925 | 2 | 0 | 19,983 | 95.14 | |
| Gravel and Brick | 282 | 536 | 4224 | 4 | 1 | 0 | 5047 | 83.69 | |
| Trees | 0 | 683 | 0 | 1874 | 0 | 0 | 2557 | 73.29 | |
| Metal Sheet | 1 | 0 | 0 | 0 | 1225 | 0 | 1226 | 99.84 | |
| Shadow | 24 | 4 | 3 | 2 | 2 | 433 | 468 | 92.52 | |
| Un-Assigned | 3658 | 2161 | 1046 | 194 | 77 | 560 | 7696 | ||
| Totals | 7926 | 22,586 | 5786 | 3001 | 1313 | 1105 | 41,717 | ||
| Producer’s agreement (%) | 49.62 | 84.18 | 73.00 | 62.45 | 93.30 | 39.19 | Overall agreement = 73.59% | ||
| Harmonic Mean (%) | 62.10 | 89.32 | 77.98 | 67.43 | 96.49 | 55.05 | Kappa Statistic = 62.10% | ||
| Reference Data | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Asphalt and Bitumen | Meadows and Soil | Gravel and Brick | Trees | Metal Sheet | Shadow | Totals | User’s Agreement (%) | ||
| Class Map | Asphalt and Bitumen | 4168 | 327 | 768 | 42 | 20 | 208 | 5533 | 75.33 |
| Meadows and Soil | 0 | 21,355 | 134 | 1106 | 6 | 10 | 22,611 | 94.45 | |
| Gravel and Brick | 0 | 493 | 4538 | 10 | 10 | 1 | 5052 | 89.83 | |
| Trees | 0 | 208 | 0 | 1751 | 0 | 5 | 1964 | 89.15 | |
| Metal Sheet | 0 | 0 | 0 | 0 | 1259 | 4 | 1263 | 99.68 | |
| Shadow | 0 | 0 | 0 | 0 | 0 | 383 | 383 | 100 | |
| Un-Assigned | 3431 | 732 | 225 | 95 | 27 | 605 | 5115 | ||
| Totals | 7599 | 23,115 | 5665 | 3004 | 1322 | 1216 | 41,921 | ||
| Producer’s agreement (%) | 54.85 | 92.39 | 80.11 | 58.29 | 95.23 | 31.50 | Overall agreement = 79.80% | ||
| Harmonic Mean (%) | 63.48 | 93.40 | 84.69 | 70.49 | 97.41 | 47.90 | Kappa Statistic = 69.30% | ||
| Reference Data | ||||||||
|---|---|---|---|---|---|---|---|---|
| Road | Grass | Trees | Roof | Dirt | Totals | User’s Agreement (%) | ||
| Class Map | Road | 5501 | 187 | 134 | 79 | 63 | 5964 | 92.24 |
| Grass | 1020 | 35,009 | 3711 | 265 | 2346 | 42,351 | 82.66 | |
| Trees | 348 | 153 | 20,654 | 961 | 208 | 22,324 | 92.52 | |
| Roof | 790 | 71 | 160 | 4727 | 108 | 5856 | 80.72 | |
| Dirt | 1481 | 369 | 309 | 126 | 3328 | 5613 | 59.29 | |
| Un-Assigned | 8325 | 722 | 1121 | 769 | 1200 | 12,137 | ||
| Totals | 17,465 | 36,511 | 26,089 | 6927 | 7253 | 94,245 | ||
| Producer’s agreement (%) | 31.50 | 95.89 | 79.17 | 68.24 | 45.88 | Overall agreement = 73.45% | ||
| Harmonic Mean (%) | 46.96 | 88.79 | 85.32 | 73.96 | 51.73 | Kappa Statistic = 64.09% | ||
| Reference Data | ||||||||
|---|---|---|---|---|---|---|---|---|
| Road | Grass | Trees | Roof | Dirt | Totals | User’s Agreement (%) | ||
| Class Map | Road | 16,476 | 4280 | 2256 | 756 | 5993 | 29,761 | 55.36 |
| Grass | 70 | 28,905 | 592 | 132 | 84 | 29,783 | 97.05 | |
| Trees | 117 | 2399 | 22,656 | 841 | 185 | 26,198 | 86.48 | |
| Roof | 738 | 76 | 315 | 4748 | 66 | 5943 | 79.89 | |
| Dirt | 0 | 53 | 25 | 289 | 594 | 961 | 61.81 | |
| Un-Assigned | 64 | 798 | 245 | 161 | 331 | 1599 | ||
| Totals | 17,465 | 36,511 | 26,089 | 6927 | 7253 | 94,245 | ||
| Producer’s agreement (%) | 94.34 | 79.17 | 86.84 | 68.54 | 8.19 | Overall agreement = 77.86% | ||
| Harmonic Mean (%) | 79.78 | 87.20 | 86.66 | 73.78 | 14.46 | Kappa Statistic = 69.95% | ||
| Reference Data | ||||||||
|---|---|---|---|---|---|---|---|---|
| Road | Grass | Trees | Roof | Dirt | Totals | User’s Agreement (%) | ||
| Class Map | Road | 16,080 | 1973 | 1215 | 282 | 444 | 19,994 | 80.42 |
| Grass | 51 | 31,226 | 1055 | 78 | 320 | 32,730 | 95.40 | |
| Trees | 136 | 1190 | 22,117 | 460 | 221 | 24,124 | 91.68 | |
| Roof | 793 | 125 | 888 | 6008 | 1157 | 8971 | 66.97 | |
| Dirt | 54 | 963 | 35 | 9 | 4752 | 5813 | 81.75 | |
| Un-Assigned | 351 | 1034 | 779 | 90 | 359 | 2613 | ||
| Totals | 17,465 | 36,511 | 26,089 | 6927 | 7253 | 94,245 | ||
| Producer’s agreement (%) | 92.07 | 85.52 | 84.78 | 86.73 | 65.52 | Overall agreement = 85.08% | ||
| Harmonic Mean (%) | 85.85 | 90.20 | 88.09 | 75.58 | 72.74 | Kappa Statistic = 79.93% | ||
| Class Harmonic Means | |||||||
|---|---|---|---|---|---|---|---|
| Road | Grass | Trees | Roof | Dirt | Kappa (%) | OA (%) | |
| Proposed: SP + QT | 87.29 | 92.97 | 89.89 | 76.06 | 81.93 | 84.43 | 88.52 |
| Full + QT | 85.85 | 90.20 | 88.09 | 75.58 | 72.74 | 79.93 | 85.08 |
| SP Global | 79.78 | 87.20 | 86.66 | 73.78 | 14.46 | 69.95 | 77.86 |
| Full Global | 46.96 | 88.79 | 85.32 | 73.96 | 51.73 | 64.09 | 73.45 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkhatib, M.Q.; Velez-Reyes, M. Improved Spatial-Spectral Superpixel Hyperspectral Unmixing. Remote Sens. 2019, 11, 2374. https://doi.org/10.3390/rs11202374
Alkhatib MQ, Velez-Reyes M. Improved Spatial-Spectral Superpixel Hyperspectral Unmixing. Remote Sensing. 2019; 11(20):2374. https://doi.org/10.3390/rs11202374
Chicago/Turabian StyleAlkhatib, Mohammed Q., and Miguel Velez-Reyes. 2019. "Improved Spatial-Spectral Superpixel Hyperspectral Unmixing" Remote Sensing 11, no. 20: 2374. https://doi.org/10.3390/rs11202374
APA StyleAlkhatib, M. Q., & Velez-Reyes, M. (2019). Improved Spatial-Spectral Superpixel Hyperspectral Unmixing. Remote Sensing, 11(20), 2374. https://doi.org/10.3390/rs11202374