A Superpixel-Based Relational Auto-Encoder for Feature Extraction of Hyperspectral Images
Abstract
:1. Introduction
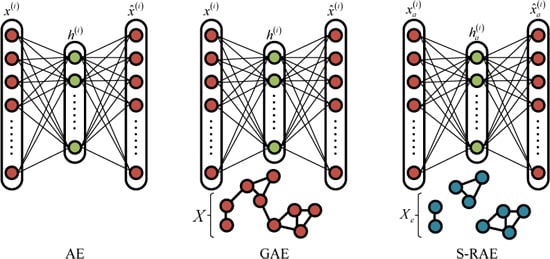
2. Graph Regularized Auto-Encoder (GAE)
3. Superpixel-Based Relational Auto-Encoder
| Algorithm 1: S-RAE. |
| Input: HSI data; Hidden size, sparsity , regular parameter , for ; Superpixel clusters number and weight coefficient for ; weighting parameter and for MS-RCAE. |
| 1. The first three principal components from PCA of HSI are reserved as the inputs of VGG16; |
| 2. Extract DSaF from the pre-trained filter banks in VGG16; |
| 3. Upsample feature maps in the last pooling layer with 4 pixels stride by bilinear interpolation operation; |
| 4. Normalize the raw spectral data and downsample with 8 pixels stride by average pooling; |
| 5. Reserve the maximum principal component of the downsampled image after PCA, and do superpixel segmentation; |
| 6. Separate the cross-region superpixels in the segmented image by connected graph method; |
7. Learning cohesive DSaF by S-RAE
|
| 8. Upsample the learned features of hidden layer to have a same scale with the input maps. |
| Output: Feature maps |
3.1. Model Establishment
3.2. Model Optimization
4. Multiscale Spectral-Spatial Feature Fusion
5. Experiments
5.1. Data Sets and Quantitative Metrics
5.2. Parameters Analysis
5.3. Stepwise Evaluation of the Proposed Strategies
5.4. Comparison with Other Feature Extraction Algorithm
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, L.; Zhang, L.; Du, B. Deep learning for remote sensing data: A technical tutorial on the state of the art. IEEE Geosci. Remote Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Jiao, L.; Zhao, J.; Yang, S.; Liu, F. Deep learning, Optimization and Recognition; Tsinghua University Press: Beijing, China, 2017. [Google Scholar]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep convolutional neural networks for hyperspectral image classification. J. Sens. 2015, 2015, 1–12. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep feature extraction and classification of hyperspectral images based on convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Xing, C.; Ma, L.; Yang, X. Stacked denoise autoencoder based feature extraction and classification for hyperspectral images. J. Sens. 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Zheng, J.; Zhao, H.; Qing, C.; Yang, Z.; Du, P.; Marshall, S. Novel segmented stacked autoencoder for effective dimensionality reduction and feature extraction in hyperspectral imaging. Neurocomputing 2016, 185, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, J.; Liu, P.; Choo, K.K.R.; Huang, F. Spectral–spatial multi-feature-based deep learning for hyperspectral remote sensing image classification. Soft Comput. 2017, 21, 213–221. [Google Scholar] [CrossRef]
- Jiao, L.; Liu, F. Wishart deep stacking network for fast POLSAR image classification. IEEE Trans. Image Process. 2016, 25, 3273–3286. [Google Scholar] [CrossRef]
- Guo, Y.; Jiao, L.; Wang, S.; Wang, S.; Liu, F. Fuzzy Sparse Autoencoder Framework for Single Image Per Person Face Recognition. IEEE Trans. Cybern. 2018, 48, 2402–2415. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, X.; Jia, X. Spectral–spatial classification of hyperspectral data based on deep belief network. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2015, 8, 2381–2392. [Google Scholar] [CrossRef]
- Zhong, P.; Gong, Z.; Li, S.; Schönlieb, C.B. Learning to diversify deep belief networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3516–3530. [Google Scholar] [CrossRef]
- Liu, F.; Jiao, L.; Hou, B.; Yang, S. POL-SAR Image Classification Based on Wishart DBN and Local Spatial Information. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3292–3308. [Google Scholar] [CrossRef]
- Zhao, Z.; Jiao, L.; Zhao, J.; Gu, J.; Zhao, J. Discriminant deep belief network for high-resolution SAR image classification. Pattern Recognit. 2017, 61, 686–701. [Google Scholar] [CrossRef]
- Zhou, Y.; Wei, Y. Learning Hierarchical Spectral-Spatial Features for Hyperspectral Image Classification. IEEE Trans. Cybern. 2017, 46, 1667–1678. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Generative Adversarial Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5046–5063. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.; Xu, X. MugNet: Deep learning for hyperspectral image classification using limited samples. ISPRS J. Photogramm. Remote Sens. 2017, 145, 108–119. [Google Scholar] [CrossRef]
- Santara, A.; Mani, K.; Hatwar, P.; Singh, A.; Garg, A.; Padia, K.; Mitra, P. BASS Net: Band-adaptive spectral–spatial feature learning neural network for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 5293–5301. [Google Scholar] [CrossRef]
- Song, W.; Li, S.; Fang, L.; Lu, T. Hyperspectral image classification with deep feature fusion network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3173–3184. [Google Scholar] [CrossRef]
- Jiao, L.; Liang, M.; Chen, H.; Yang, S.; Liu, H.; Cao, X. Deep fully convolutional network-based spatial distribution prediction for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 5585–5599. [Google Scholar] [CrossRef]
- Mei, S.; Ji, J.; Hou, J.; Li, X.; Du, Q. Learning sensor-specific spatial–spectral features of hyperspectral images via convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4520–4533. [Google Scholar] [CrossRef]
- Lin, Z.; Chen, Y.; Zhao, X.; Wang, G. Spectral–spatial classification of hyperspectral image using autoencoders. In Proceedings of the International Conference on Information, Communications & Signal Processing (ICICS), Tainan, Taiwan, 10–13 December 2013; pp. 1–5. [Google Scholar]
- Zhang, X.; Liang, Y.; Li, C.; Huyan, N.; Jiao, L.; Zhou, H. Recursive Autoencoders-Based Unsupervised Feature Learning for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1928–1932. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Zhou, F.; Xu, L.; Meng, D.; Xu, Z.; Paisley, J. Hyperspectral image classification with Markov random fields and a convolutional neural network. IEEE Trans. Image Process. 2018, 27, 2354–2367. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, Y.Q.; Chan, J.C.W. Learning and Transferring Deep Joint Spectral–Spatial Features for Hyperspectral Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4729–4742. [Google Scholar] [CrossRef]
- Hu, F.; Xia, G.S.; Hu, J.; Zhang, L. Transferring deep convolutional neural networks for the scene classification of high-resolution remote sensing imagery. Remote Sens. 2015, 7, 14680–14707. [Google Scholar] [CrossRef]
- Hastad, J.; Goldmann, M. On the power of small-depth threshold circuits. Comput. Complex. 1991, 1, 113–129. [Google Scholar] [CrossRef]
- Li, L.; Zhang, T.; Shan, C.; Liu, Z. Deep Learning: Mastering Convolutional Neural Networks from Beginner; China Machine Press: Beijing, China, 2018. [Google Scholar]
- Makantasis, K.; Karantzalos, K.; Doulamis, A.; Doulamis, N. Deep supervised learning for hyperspectral data classification through convolutional neural networks. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 4959–4962. [Google Scholar]
- Yue, J.; Zhao, W.; Mao, S.; Liu, H. Spectral—Spatial classification of hyperspectral images using deep convolutional neural networks. Remote Sens. Lett. 2015, 6, 468–477. [Google Scholar] [CrossRef]
- Lee, H.; Kwon, H. Contextual deep CNN based hyperspectral classification. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 3322–3325. [Google Scholar]
- Liang, M.; Jiao, L.; Yang, S.; Liu, F.; Hou, B.; Chen, H. Deep Multiscale Spectral-Spatial Feature Fusion for Hyperspectral Images Classification. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2018, 11, 2911–2924. [Google Scholar] [CrossRef]
- Maaten, L.V.D.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Research 2008, 9, 2579–2605. [Google Scholar]
- Liao, Y.; Wang, Y.; Liu, Y. Graph regularized auto-encoders for image representation. IEEE Trans. Image Process. 2017, 26, 2839–2852. [Google Scholar] [CrossRef]
- Liu, M.Y.; Tuzel, O.; Ramalingam, S.; Chellappa, R. Entropy rate superpixel segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Providence, RI, USA, 20–25 June 2011; pp. 2097–2104. [Google Scholar]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Li, S.; Benediktsson, J.A. Feature extraction of hyperspectral images with image fusion and recursive filtering. IEEE Trans. Geosci. Remote Sens. 2014, 52, 3742–3752. [Google Scholar] [CrossRef]
- Kang, X.; Li, S.; Fang, L.; Benediktsson, J.A. Intrinsic image decomposition for feature extraction of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2015, 53, 2241–2253. [Google Scholar] [CrossRef]















| Class | # Samples | Classification Methods | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | SVM | IID | RF | J-SAE | GF-FSAE | 3D-CNN | MSFE | DMSF | MS-RCAE | |
| 1 | 3 | 43 | 34.65 | 97.98 | 94.53 | 97.56 | 61.74 | 60.98 | 95.47 | 97.44 | 96.51 |
| 2 | 72 | 1356 | 65.38 | 83.38 | 92.90 | 85.68 | 67.19 | 78.60 | 88.84 | 96.23 | 96.39 |
| 3 | 42 | 788 | 43.87 | 89.75 | 93.05 | 90.50 | 74.00 | 87.42 | 93.78 | 96.55 | 96.51 |
| 4 | 12 | 225 | 34.64 | 86.50 | 90.07 | 68.22 | 58.69 | 88.32 | 92.87 | 96.00 | 96.49 |
| 5 | 25 | 458 | 81.08 | 94.11 | 92.72 | 78.98 | 89.20 | 80.60 | 92.31 | 93.49 | 94.65 |
| 6 | 38 | 692 | 93.16 | 97.64 | 99.34 | 95.11 | 98.37 | 92.98 | 98.89 | 99.62 | 99.65 |
| 7 | 2 | 26 | 65.19 | 96.28 | 98.46 | 60.00 | 51.54 | 68.00 | 96.54 | 99.62 | 99.62 |
| 8 | 25 | 453 | 95.20 | 100 | 99.67 | 100 | 99.28 | 95.57 | 99.22 | 100 | 100 |
| 9 | 2 | 18 | 34.17 | 99.63 | 88.89 | 44.44 | 47.22 | 77.78 | 100 | 95.00 | 96.67 |
| 10 | 49 | 923 | 61.16 | 84.07 | 92.38 | 83.43 | 75.14 | 76.91 | 92.32 | 94.54 | 94.43 |
| 11 | 124 | 2331 | 78.29 | 86.72 | 96.33 | 95.24 | 60.53 | 85.42 | 98.72 | 98.82 | 98.79 |
| 12 | 31 | 562 | 44.77 | 81.49 | 91.93 | 91.17 | 65.23 | 82.52 | 92.78 | 94.11 | 94.56 |
| 13 | 11 | 194 | 97.40 | 98.97 | 99.10 | 91.30 | 94.48 | 96.20 | 98.69 | 96.39 | 98.56 |
| 14 | 65 | 1200 | 95.74 | 99.20 | 98.28 | 97.01 | 99.31 | 99.30 | 99.95 | 99.06 | 99.28 |
| 15 | 19 | 367 | 42.33 | 89.52 | 93.96 | 95.98 | 90.89 | 89.94 | 99.46 | 98.69 | 98.96 |
| 16 | 5 | 88 | 85.34 | 90.04 | 98.30 | 86.75 | 81.42 | 85.54 | 93.81 | 92.84 | 98.41 |
| AA (%) | 65.77 | 92.21 | 94.99 | 85.09 | 75.89 | 84.13 | 95.85 | 96.78 | 97.47 | ||
| OA (%) | 72.04 | 89.70 | 95.22 | 90.89 | 76.77 | 86.43 | 95.71 | 97.30 | 97.53 | ||
| Kappa | 0.6775 | 0.8828 | 0.9455 | 0.8960 | 0.7442 | 0.8450 | 0.9510 | 0.9693 | 0.9718 | ||
| Class | # Samples | Classification Methods | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | SVM | IID | RF | J-SAE | GF-FSAE | 3D-CNN | MSFE | DMSF | MS-RCAE | |
| 1 | 10 | 6621 | 62.75 | 80.40 | 65.55 | 58.25 | 75.60 | 57.48 | 87.67 | 92.80 | 95.15 |
| 2 | 10 | 18,639 | 58.75 | 88.02 | 84.34 | 86.19 | 76.36 | 87.80 | 87.38 | 91.06 | 94.17 |
| 3 | 10 | 2089 | 46.45 | 94.25 | 82.93 | 58.34 | 83.83 | 53.20 | 97.87 | 97.95 | 98.95 |
| 4 | 10 | 3054 | 91.41 | 92.49 | 77.41 | 95.78 | 95.34 | 89.19 | 88.06 | 91.90 | 94.30 |
| 5 | 10 | 1335 | 99.77 | 99.98 | 99.19 | 100 | 94.84 | 95.09 | 99.98 | 99.62 | 99.81 |
| 6 | 10 | 5019 | 59.20 | 98.28 | 93.47 | 69.99 | 88.01 | 75.70 | 95.36 | 97.27 | 98.44 |
| 7 | 10 | 1320 | 88.18 | 99.01 | 88.24 | 95.15 | 85.77 | 89.08 | 99.50 | 98.48 | 99.37 |
| 8 | 10 | 3672 | 73.23 | 86.14 | 67.62 | 57.75 | 81.68 | 86.95 | 93.47 | 95.59 | 95.70 |
| 9 | 10 | 937 | 99.57 | 84.23 | 66.90 | 96.95 | 93.78 | 92.88 | 98.69 | 97.39 | 97.75 |
| AA (%) | 75.48 | 91.42 | 80.63 | 79.82 | 86.14 | 80.82 | 94.22 | 95.78 | 97.07 | ||
| OA (%) | 65.59 | 89.14 | 80.70 | 75.66 | 81.05 | 80.39 | 90.51 | 93.52 | 95.63 | ||
| Kappa | 0.5765 | 0.8599 | 0.7513 | 0.6823 | 0.7603 | 0.7454 | 0.8781 | 0.9165 | 0.9435 | ||
| Class | # Samples | Classification Methods | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | SVM | IID | RF | J-SAE | GF-FSAE | 3D-CNN | MSFE | DMSF | MS-RCAE | |
| 1 | 10 | 1999 | 96.81 | 100 | 100 | 100 | 97.51 | 78.68 | 97.97 | 97.09 | 99.99 |
| 2 | 10 | 3716 | 91.91 | 99.95 | 97.87 | 100 | 99.04 | 75.50 | 97.40 | 100 | 100 |
| 3 | 10 | 1966 | 88.71 | 100 | 99.99 | 94.79 | 100 | 97.09 | 99.63 | 97.58 | 98.55 |
| 4 | 10 | 1384 | 99.28 | 99.29 | 98.94 | 98.84 | 99.41 | 99.85 | 99.45 | 98.79 | 99.65 |
| 5 | 10 | 2668 | 95.97 | 98.88 | 95.93 | 96.61 | 97.90 | 97.07 | 97.61 | 94.39 | 95.34 |
| 6 | 10 | 3949 | 99.74 | 99.86 | 99.60 | 100 | 99.84 | 99.85 | 99.81 | 99.79 | 99.95 |
| 7 | 10 | 3569 | 99.51 | 99.86 | 99.02 | 95.98 | 99.80 | 99.72 | 99.94 | 99.10 | 99.85 |
| 8 | 10 | 11,261 | 61.09 | 94.97 | 87.62 | 68.59 | 84.62 | 29.83 | 92.50 | 85.66 | 93.30 |
| 9 | 10 | 6193 | 97.85 | 98.89 | 99.99 | 98.53 | 99.81 | 95.05 | 98.31 | 99.63 | 99.69 |
| 10 | 10 | 3268 | 76.73 | 96.84 | 98.40 | 88.80 | 89.10 | 92.33 | 92.21 | 95.64 | 96.96 |
| 11 | 10 | 1058 | 91.55 | 99.73 | 96.61 | 99.14 | 95.20 | 100 | 97.37 | 99.15 | 99.89 |
| 12 | 10 | 1917 | 97.52 | 100 | 97.13 | 99.11 | 99.13 | 99.42 | 99.18 | 97.22 | 99.42 |
| 13 | 10 | 906 | 95.27 | 98.37 | 97.13 | 98.21 | 92.23 | 96.99 | 96.69 | 98.17 | 99.77 |
| 14 | 10 | 1060 | 91.30 | 96.88 | 96.75 | 99.05 | 94.08 | 100 | 94.52 | 98.36 | 99.28 |
| 15 | 10 | 7258 | 58.03 | 93.29 | 97.00 | 53.08 | 91.27 | 86.81 | 96.15 | 90.13 | 92.12 |
| 16 | 10 | 1797 | 91.92 | 99.75 | 95.77 | 98.27 | 97.02 | 94.18 | 99.53 | 99.64 | 99.99 |
| AA (%) | 89.57 | 98.53 | 97.36 | 93.04 | 95.98 | 90.15 | 97.39 | 96.90 | 98.36 | ||
| OA (%) | 82.45 | 97.53 | 96.02 | 85.43 | 94.15 | 79.49 | 96.57 | 94.61 | 96.98 | ||
| Kappa | 0.8052 | 0.9725 | 0.9558 | 0.8379 | 0.9350 | 0.7746 | 0.9619 | 0.9401 | 0.9663 | ||
| Class | # Samples | Classification Methods | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | SVM | IID | RF | J-SAE | GF-FSAE | 3D-CNN | MSFE | DMSF | MS-RCAE | |
| 1 | 10 | 751 | 81.91 | 73.88 | 83.93 | 93.66 | 86.60 | 91.50 | 94.10 | 96.36 | 99.19 |
| 2 | 10 | 233 | 74.59 | 74.06 | 67.30 | 73.99 | 83.67 | 100 | 93.24 | 98.37 | 99.10 |
| 3 | 10 | 246 | 82.99 | 95.77 | 99.51 | 84.75 | 72.09 | 85.59 | 98.70 | 98.50 | 99.88 |
| 4 | 10 | 242 | 40.21 | 74.71 | 77.87 | 39.66 | 60.56 | 60.34 | 97.62 | 94.55 | 95.87 |
| 5 | 10 | 151 | 40.36 | 82.65 | 97.15 | 58.16 | 52.45 | 100 | 86.49 | 98.08 | 98.08 |
| 6 | 10 | 219 | 51.16 | 99.82 | 86.67 | 77.03 | 30.82 | 94.26 | 99.82 | 100 | 100 |
| 7 | 10 | 95 | 78.53 | 100 | 99.32 | 100 | 69.63 | 100 | 100 | 100 | 100 |
| 8 | 10 | 421 | 73.97 | 95.49 | 95.12 | 77.62 | 89.26 | 85.89 | 93.57 | 94.21 | 98.26 |
| 9 | 10 | 510 | 81.14 | 83.76 | 89.73 | 90.60 | 86.62 | 73.80 | 94.63 | 98.02 | 98.35 |
| 10 | 10 | 394 | 79.12 | 95.30 | 94.43 | 88.02 | 95.75 | 99.48 | 88.85 | 100 | 100 |
| 11 | 10 | 409 | 91.49 | 99.58 | 99.27 | 97.99 | 98.22 | 93.23 | 96.32 | 100 | 100 |
| 12 | 10 | 493 | 86.90 | 95.43 | 91.45 | 92.55 | 83.09 | 83.23 | 96.98 | 97.63 | 100 |
| 13 | 10 | 917 | 99.91 | 99.99 | 100 | 100 | 99.44 | 100 | 100 | 100 | 100 |
| AA (%) | 74.02 | 90.03 | 90.90 | 82.62 | 77.55 | 89.79 | 95.41 | 98.13 | 99.12 | ||
| OA (%) | 80.58 | 90.17 | 91.62 | 87.54 | 84.63 | 89.90 | 95.71 | 98.10 | 99.26 | ||
| Kappa | 0.7840 | 0.8909 | 0.9067 | 0.8609 | 0.8283 | 0.8876 | 0.9523 | 0.9788 | 0.9918 | ||
| Datasets\Methods | MS-RCAE (s) | J-SAE (s) | GF-FSAE (s) | DMSF (s) | 3D-CNN (m) | MSFE (s) |
|---|---|---|---|---|---|---|
| Indian Pines | 163.45 | 310.94 | 247.69 | 132.12 | 41.24 | 0.56 |
| University of Pavia | 146.99 | 274.30 | 298.48 | 127.76 | 23.71 | 2.30 |
| Salinas | 194.69 | 305.23 | 282.85 | 160.43 | 50.53 | 1.79 |
| KSC | 291.70 | 430.78 | 291.89 | 225.71 | 44.51 | 3.72 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, M.; Jiao, L.; Meng, Z. A Superpixel-Based Relational Auto-Encoder for Feature Extraction of Hyperspectral Images. Remote Sens. 2019, 11, 2454. https://doi.org/10.3390/rs11202454
Liang M, Jiao L, Meng Z. A Superpixel-Based Relational Auto-Encoder for Feature Extraction of Hyperspectral Images. Remote Sensing. 2019; 11(20):2454. https://doi.org/10.3390/rs11202454
Chicago/Turabian StyleLiang, Miaomiao, Licheng Jiao, and Zhe Meng. 2019. "A Superpixel-Based Relational Auto-Encoder for Feature Extraction of Hyperspectral Images" Remote Sensing 11, no. 20: 2454. https://doi.org/10.3390/rs11202454
APA StyleLiang, M., Jiao, L., & Meng, Z. (2019). A Superpixel-Based Relational Auto-Encoder for Feature Extraction of Hyperspectral Images. Remote Sensing, 11(20), 2454. https://doi.org/10.3390/rs11202454