1. Introduction
The science and technology of remote sensing has reached the era of big data [
1]. This imposes a paradigm change in the way remote sensing data is processed to extract information for a variety of environmental and societal applications. The proliferation of constellations of compact satellites adds another level of challenge to processing remote sensing imagery [
2]. The extraction of information from this type of data, regardless of the application category (e.g., global climate change, urban planning, land-use and land-cover (LULC) monitoring), can be classified as big remote sensing data, while simultaneously meeting volume, variety, and data growth rates [
3]. The challenges involved in this process have led to the emergence of cloud-based platforms, specifically for remote sensing, that can perform planetary-scale analysis of massive amounts of data [
4,
5,
6,
7].
The combination of these factors, i.e., the high availability of satellite data and adequate analysis platforms, has enabled the emergence of terrestrial coverage mappings on global [
8,
9,
10], continental [
11,
12,
13], and national [
14,
15,
16] scales, which are useful for supporting decision-making and broader policy objectives [
17]. In general, these mappings were produced with supervised classifications using conventional machine learning algorithms (e.g., random forest, classification and regression tree-CART) implemented on platforms with thousands of processors capable of handling communication, parallelism, and distributed computing problems [
18].
Although the operation of these algorithms for large geographic regions uses cloud-based platforms, the classification approaches that are implemented generally use a step of feature engineering to handle and reduce the dimensionality of the input satellite data. In addition to allowing faster and computationally more efficient execution, this step provides a better understanding of the features that are most relevant to the training process and that most often impacts the accuracy of the classification result [
19,
20]. Typically, feature engineering applied to remote sensing depends on human experience and domain knowledge, and is time-consuming as it needs to consider a lot of complex factors simultaneously (e.g., inter-class similarity, intra-class variability, atmospheric conditions [
21]). Furthermore, as we enter a new era of big remote sensing data, this step becomes more arduous and challenging.
In this scenario, deep learning algorithms (e.g., convolutional neural networks, recurrent neural networks, semantic segmentation [
22]) may be more appropriate as they are able to construct and select the most relevant features of raw data automatically, without human interference, which can result in better and more accurate mapping products [
23]. In fact, the deep learning approaches have proven their effectiveness in many application domains, dramatically improving speech recognition, visual object recognition, health diagnosis, and others applications [
24]. However, most studies that apply deep learning approaches to remote sensing data are restricted to the use of publicly available benchmark image datasets that involve pre-processed images organized under ideal conditions and have been previously labeled [
25,
26,
27], such that research focuses on the comparative analysis of accuracy gains. Because of that, practical applications of deep learning algorithms for land-use and land-cover mapping, operationally and for large geographic regions, require further research investigations [
22,
28].
On the other hand, deep learning algorithms generally require substantially more training data than conventional machine learning supervised classifications [
24]. Acquiring training data is challenging due to the high costs and difficulties involved in obtaining field data, especially for past years [
29], as well as due to the subjectivity and time-consuming work involved in the visual interpretation of remote sensing images [
30]. In this sense, the feature engineering exemption from deep learning is likely to be offset by a greater need for samples, a trade-off that should be considered when designing national, continental, and global mapping products.
Another trade-off that needs to be considered when using deep learning algorithms concerns the increased need for computational processing, which is usually met by using graphical processing units (GPUs) [
24]. Although there are initiatives that seek to advance GPU satellite data processing [
31,
32], existing solutions are not as operational as cloud-based platforms that make exclusive use of CPUs, such as the Google Earth Engine [
5]. From this perspective, we can state that the next generation of land surface mappings involves an adequate understanding of the benefits and trade-offs between conventional (e.g., random forest, CART, support vector machine-SVM) and cutting-edge (e.g., Google LeNet, U-Net, generative adversarial network-GAN) machine learning algorithms when applied to remote sensing, as well as a proper definition and evaluation of operational strategies that is capable of promoting better information extraction.
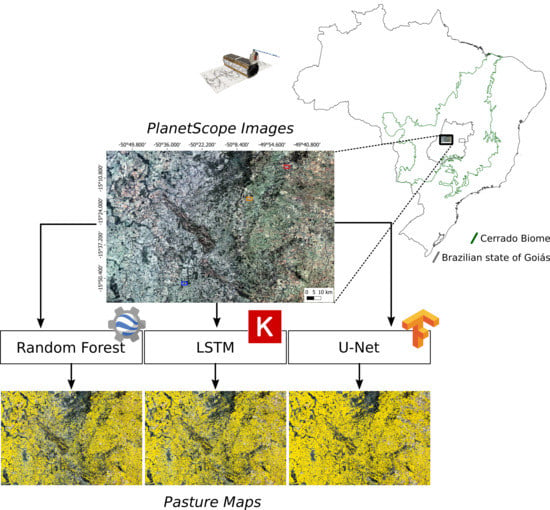
Aiming to contribute to the next generation of land-cover and land-use maps, we evaluated the performance of random forest, long short-term memory (LSTM), and U-Net algorithms, which were applied to a large dataset of PlanetScope satellite imagery, and implemented in the Google Earth Engine and in a local infrastructure using Tensorflow. Additionally, we present a framework regarding pre-processing, the preparation of training data, feature engineering, and accuracy assessment, and discuss the challenges faced in implementing such novel methods to mapping very large areas.
2. Materials and Methods
We focused the implementation of machine learning and deep learning algorithms on the mapping of pasture areas, one of the most complex LULC classes to map [
33], using PlanetScope imagery obtained with the Planet Dove constellation. With over 130 commercial CubeSats in orbit, the Planet constellation enables the monitoring of the entire terrestrial surface with a daily frequency and a spatial resolution of 3–5m, producing a very large data volume, which fits into the realm of big remote sensing data, despite not presenting the same radiometric quality and calibration precision of government satellites (e.g., Sentinel 2 and Landsat 8) [
2].
Specifically, we used thousands of PlanetScope images acquired over a one-year period to produce 12 monthly mosaics over a study area located in central Brazil [
34]. Based on these mosaics, a specific training dataset for mapping pasture areas in the region was produced by considering aspects such as the local variability of LULC classes and potential spectral confusion. The monthly mosaics and training set were used to build supervised classification models using the random forest [
35], LSTM [
36], and U-Net [
37] algorithms. In relation to the random forest, a conventional machine learning approach, a feature engineering step was performed, aiming to improve its performance in the detection of pasture areas. For LSTM and U-Net, both of which are deep learning approaches, the model building considered only raw data (i.e., the monthly mosaics). At the end of the process, the three resulting mappings were evaluated by considering the validation data, previously separated from the training data, and data obtained via fieldwork in the study area (considered as test data). The datasets and methodological steps used in this study are presented in
Figure 1 and are detailed in the following sections.
2.1. Pre-processing of PlanetScope Images
Our study area was a region of ≈18,000 km
2 in area, delimited by the topographic chart SD-22-ZC [
38], and located in the central portion of the Cerrado biome in the Brazilian state of Goiás (
Figure 2). This region was evaluated using 36,962 PlanetScope images (i.e., four-band PlanetScope Scenes), acquired between August 2017 and July 2018. We used this large imagery dataset to produce 12 monthly mosaics, which were screened for clouds and/or cloud-shadows contamination. Cloud and cloud-shadow pixels were identified using histogram slicing, which used empirically defined spectral thresholds, along with a frequency filter that prioritized pixels with low temporal stability, and consequently, had a greater chance of being associated with clouds and/or cloud shadows. The mosaicking process considered a top-of-atmosphere correction, based on Landsat reference values, and the normalization of the PlanetScope images with less than 20% cloud coverage in order to minimize the amount of noise in the final mosaic. The normalization process was based on a histogram adjustment, using an annual median mosaic (produced with only cloudless images, i.e., cloud-cover <1%, obtained over 12 months) as a reference. The histogram adjustments were implemented using statistical regressions (via a random forest) between the cumulative distributions of the PlanetScope images and the reference mosaic, band by band. Finally, the daily PlanetScope images were stacked and reduced using the median to generate the monthly mosaics. All of this was implemented on the Google Earth Engine platform, and the final mosaics, resampled to 4 m, can be accessed at
https://code.earthengine.google.com/bf9fb20125dede14ab0457490a5a8f42 [
34]
2.2. Training, Validation, and Test Data
Based on the PlanetScope of the study area, we collected pasture and non-pasture samples in order to build the training, validation, and test datasets. Considering the monthly mosaics, as well as having prior knowledge of the region, image interpretation experts collected 1326 regular and homogeneous polygons (i.e., with pixels of only one LULC class), well-distributed throughout the study area and covering, as much as possible, all the LULC class variability (
Figure 2a). For the pasture class, we collected polygons of “regular pasture,” “wet pasture,” “pasture with bare soil,” and “pasture with shrubs”; for the not-pasture class, we collected polygons of “crop,” “deforestation,” “forest formation,” “other land-use,” “planted forest,” “sand bank,” “savannah formation,” “urban area,” and “water.” These polygons were used to generate point and segment samples, which were used for the classifier training and validation. For the test set, fieldwork data obtained from the study area were used.
2.2.1. Sampling Points
Aiming to minimize the spatial autocorrelation of the point samples, a minimum distance between samples located within the same polygon was established, which varied according to the LULC classes defined by the interpreters. These distances were calculated according to Equation (1) (defined using the relation between the number of pixels and the area of a perfect square polygon), which considered the same number of samples for pasture subclasses and non-pasture subclasses (
Table 1):
where for each LULC class:
d is the minimum class distance,
N is the total of pixels in the class,
n is the number of desired samples in a given class, and
s is the pixel size.
At the end of the process, 30,000 point samples were generated, with balanced amounts between pasture and non-pasture (
Figure 3) that was representative of the study area and large enough to accommodate the increasing number of data dimensions in order to meet best training practices for supervised classifiers [
19]. Considering the generated samples, 90% of the points were used for training and 10% were used for validation of the random forest and LSTM algorithms (the utilized datasets are available at:
https://www.lapig.iesa.ufg.br/drive/index.php/s/DE3vjlUugyhEOwD).
2.2.2. Sampling Segments
The generation of segment samples, necessary for the training of semantic segmentation algorithms (e.g., U-Net), considered a regular grid over the study area that was composed of ≈14,300 cells with a cell size defined by a function of the established input for the U-Net in this study (i.e., 286 × 286 pixels;
Figure 3). Referring to this regular grid, all cells containing at least one polygon collected by the experts were selected, resulting in 1440 cells. The area delimited by these cells (i.e., ≈1800 km
2) was segmented via the mean shift algorithm and the Orfeo Toolbox/Monteverdi framework [
39], and classified via visual interpretation in the Quantum GIS software [
40], using the PlanetScope Mosaic of November 2017 as a reference.
In order to generate pasture segments capable of aggregating visible objects in very high-resolution images (e.g., isolated trees, patches of bare soil), the following parameters were defined for the mean shift algorithm: tile size of 286 × 286, spatial radius of 24, range radius of 128, and a minimum segment size of 4 (all in pixel units). The choice of the November 2017 mosaic was justified by the greater vegetative vigor of the pasture areas due to the beginning of the rainy season, and the presence of bare soil in the agricultural areas due to the beginning of the cropping cycle, factors that generally facilitate classification based on segmentation and visual interpretation. This process resulted in ≈937,000 segments, with 36% of the segments classified as pasture, which were converted to a raster format compatible with the PlanetScope mosaics (i.e., same pixel size and geographic coordinate system;
Figure 3). Considering the total number of selected cells and their respective segments in the raster format, 90% of the cells were used for training and 10% were used for validation of the U-Net algorithm (the utilized datasets are available at
https://www.lapig. iesa.ufg.br/drive/index.php/s/DE3vjlUugyhEOwD).
2.2.3. Field Data
Field data were obtained for three different periods between 2017 and 2018, comprising pasture and non-pasture regions. This study considered a total of 262 samples containing geographic coordinates, LULC classes (i.e., “regular pasture,” “well-managed pasture,” “pasture with bare soil,” “pasture with shrubs,” “forest formation,” “savannah formation,” “crop,” “sugarcane,” “planted forests,” “urban area,” and “water”), area characterization information, and field pictures. These data, relatively concentrated in the southwestern portion of the study area (
Figure 2b), were used as test data in the accuracy analysis of the produced mappings.
2.3. Random Forest Classification
The random forest training utilized a set of texture-spectral and temporal metrics generated from the monthly mosaics and 27,000 point samples to build 100 statistical decision trees. The growth of each tree considered 50% of the training samples as a bag fraction, six random spectral metrics to be selected in the definition of the best split node (Mtry), and one sample as a minimum leaf population of a terminal node. Considering its low sensitivity to overfitting, the most common recommendation for a random forest is to set a large number of trees and the square root of the feature space size as the number of variables for splitting [
19]. The set of metrics was defined using a feature engineering strategy, which evaluated different time windows and opted for the one that produced the best pasture mapping of the study area according to the validation samples. All of these processing steps were implemented on the Google Earth Engine platform and are available at:
https://code. earthengine.google.com/2bf9aebba03eac0d1e55d79731847cff.
Feature Engineering
The feature engineering used all the spectral information available on the PlanetScope images (i.e., the blue, green, red, and near-infrared bands), as well as the normalized difference vegetation index (NDVI) [
41], the photochemical reflectance index (PRI) [
42], and the normalized difference water index (NDWI) [
43] to generate a set of spectral-temporal metrics, an approach that is already consolidated in pasture operational mappings with Landsat [
15] and MODIS [
44] satellite data. The temporal aspect was incorporated via statistical operations that reduced multiple monthly mosaics to a single spectral value in order to capture the seasonal aspects of pasture and non-pasture pixels. This process used the median and three other statistical reducers derived from the standard deviation (i.e., +2σ to obtain maximum values, −2σ to obtain minimum values, and 4σ to obtain the amplitude of the spectral variation), yielding 28 spectral-temporal metrics overall.
Aiming to improve the classification efficiency of very high spatial resolution pixels, the feature engineering also considered texture metrics capable of capturing spatial and topological aspects. We applied these metrics on the green, red, and near-infrared spectral bands using the gray-level co-occurrence matrix (GLCM) method and a 3 × 3 pixel spatial window [
45]. Based on the assessment of the GLCM result, we opted for the use of contrast, second angular momentum, and entropy metrics, which are capable of highlighting transition zones, homogeneous regions, and the degree of disorder in the land-use and land-cover classes in the landscape, respectively. The application of these metrics was done for multiple monthly mosaics, and at the end of the process, a median reducer was used in each spectral band to generate nine texture metrics.
The feature space, composed of these 37 variables (i.e., 28 spectral-temporal metrics and 9 texture metrics), was generated for 9 distinct time windows, considering only one month (i.e., November and March); every quarter (i.e., August–October, November–January, February–April, and May–July); two semesters (i.e., August–January and February–July); and 12 months (i.e., August–July) of the analyzed period. Specifically, for the one-month classifications, 16 metrics were considered (i.e., four spectral bands, three spectral indices, and nine texture metrics). This process generated nine feature spaces, and consequently, nine classification results, which were evaluated through the validation set derived from the point samples. The result that presented the highest F1 score, which was a metric capable of simultaneously considering omission and commission errors [
46], was considered as the best mapping produced with the random forest, which was used in the accuracy analysis and in the comparison with the other pasture mappings (i.e., produced via deep learning approaches).
2.4. Long Short-Term Memory Classification
The implementation of the LSTM [
36] classification approach considered a neural network architecture that simultaneously analyzed the spectral, spatial, and temporal dimensions of all monthly PlanetScope mosaics (
Figure 4). The LSTM bidirectional layer processed the spectral and temporal dimensions by analyzing the two-way time series (i.e., from beginning to end, and from end to beginning) in order to seek seasonal patterns capable of separating pasture and non-pasture pixels. The first 3D convolution layer used 256 kernels with a 3 × 3 × 5 size (i.e., a 3 × 3 spatial window over five months) to process the PlanetScope monthly mosaics, maintaining the same time-series size as the output. The second 3D convolution layer used a valid padding strategy to transform the 3D time-series into a 2D time-series, which was later concatenated with the output of the LSTM layer and processed using a set of fully-connected layers. Prior to concatenation, a batch normalization operations [
47] was applied, and between the fully-connected layers, a dropout rate of 0.3 [
48] was applied, which generally decreased the chance of overfitting during model training. Because it is a binary classification, the output layer used the sigmoid activation function, while the other layers used the ReLu function.
The training of the neural network occurred for 100 epochs with a learning rate of 5 × 10
−5, the binary cross-entropy as a cost function, and the Nadam optimizer, which incorporates the nesterov momentum in the Adam optimizer [
49]. In order to make possible the comparison between the results produced by this approach and the random forest, we used the same set of training and validation with, respectively, 27,000 and 3000 point samples, with a mini-batch of size 16. Finally, the trained model was used to classify all the pixels in the study area and produce a pasture map for the region.
All the processing steps were performed on a local server with an Intel Xeon E5-2620 v2 processor, 48 GB of memory, and two GPU cards (NVIDIA Titan-X and NVIDIA 1080ti). The neural network was developed in Keras using Tensorflow, with the LSTM implementation in NVIDIA cuDNN [
50,
51], while the other operations involving the PlanetScope mosaics (e.g., access and preparation of the time-series) made use ofGeospatial Data Abstraction Library-GDAL and NumPy libraries [
52]. The source code used by this classification approach can be accessed at
https://github.com/NexGenMap/dl-time-series.
2.5. U-Net Classification
We used the original U-Net architecture [
37] with some modifications to consider the temporal dimension and a set of more recent hyperparameters (
Figure 5). The incorporation of time occurred by stacking the spectral bands of the March and November monthly mosaics. The use of the November mosaic was expected as it was used in the generation of the segment samples, while the option for the March mosaic was justified by its relevance in the rainy season. In March, the pastures generally show a higher vegetative vigor and some agricultural areas are at the peak of cultivation, showing complementary aspects to those observed in November, which can therefore contribute to a better performance of the U-Net regarding pasture discrimination.
With regard to the new hyperparameters, all convolutional layers considered the batch normalization operation [
47] and an L2 regularization of 0.5 [
53], while specifically for the penultimate layer, a dropout rate of 0.5 was utilized [
49]. The cost function used was the intersection-over-union, which is generally better suited to binary semantic segmentation problems [
54], and the optimizer chosen was Nadam [
50]. We chose this cost function to minimize the impact of the class imbalance in the segment samples, which is a common issue of binary semantic segmentation problems, with the aim of enabling a fair comparison with the other classification approaches (i.e., random forest and LSTM-based architecture) that used balanced point samples (
Table 1). With a learning rate of 5 × 10
−5 and a 32-size mini-batch, the neural network training occurred for 100 epochs and considered all segment samples in raster format to generate the input chips (i.e., a set of pixels with a regular squared size), which were submitted to two traditional data augmentation strategies, three rotations (i.e., 90°, 180°, and 270°), and a flip operation (i.e., left–right [
55]).
Originally, the U-net was designed to receive, as an input, an image larger than its output layer, and for the sake of this work, this feature was maintained in the training of the model (i.e., 286 × 286 pixels input chips and 100 × 100 pixels output). By considering pixels adjacent to and external to the classification area, this strategy can yield more spatially consistent mapping results. In this sense, the trained model was applied throughout the study area by using a sequential scanning approach and a floating window of 100 × 100 pixels. For the classification of regions at the borders of the study area (e.g., upper-left and lower-right corners), the missing pixels needed for the input chip were artificially filled with a reflect technique implemented by the SciPy library [
56]. The other processing steps were developed with the Tensorflow [
51] and GDAL [
52] libraries, while the training and classification were performed in the same hardware environment used by the LSTM. The source code used by this classification approach can be accessed at
https://github.com/NexGenMap/dl-semantic-segmentation.
2.6. Accuracy Analysis
The accuracy analysis was performed for the three pasture mappings produced by considering the validation samples previously separated from the training samples and the test samples obtained in the field. For both sample sets, we calculated the producer (i.e., recall) and user (i.e., precision) accuracies, the F1 score (i.e., a metric calculated as a function of both the producer and user accuracies) [
56], and the population estimate of the error matrix [
30], calculated according to the pasture area mapped by each classification approach.
4. Discussion
Given the possibility of applying conventional and state-of-the-art machine learning algorithms to large volumes of satellite data, this study used different classification approaches to produce and evaluate three pasture maps for an area of 18,000 km2. The use of images obtained by the PlanetScope constellation required a sampling strategy compatible with very high spatial resolution data that was representative for the study area. Given the classification results obtained, we can assume that the chosen strategy was adequate since the mapping products achieved F1 scores greater than 95% for the validation data and 82% for the test data. We believe that part of the success of these results can be attributed to the representativeness of the LULC classes and subclasses, defined by experts with knowledge of the study area, which eventually allowed for the algorithms to generate more generic and comprehensive classification rules to separate between pasture and non-pasture pixels. In this sense, we assume that the classes and subclasses identified by visual interpretation could also be differentiated by some machine learning approach.
On the other hand, the need for knowledge regarding the mapping region for the definition of classes and subclasses can be considered a limitation of this sample collection strategy, especially with regard to large-scale mappings. The use of an unsupervised classification approach [
57] could be a possible solution to this lack of knowledge, allowing specialists to define regular and homogeneous polygons within a set of automatically defined classes. Another possible modification would be to include samples with some level of LULC class mixing, as conventional classification algorithms can handle different levels of heterogeneity in training samples, although this practice does not bring significant accuracy gains [
58,
59].
Specifically in terms of the feature engineering, the relationship between the best classification results with random forest and the monthly mosaics obtained in the rainy season was an interesting finding. This was because, in general, the images have a high cloud coverage during this period, and we can say that the high temporal resolution of the PlanetScope data was crucial for obtaining full coverage and therefore cloud-free mosaics for every month. Regarding the spectral resolution, we recognize that despite the low radiometric quality and lack of calibration of the PlanetScope constellation, a common characteristic in CubeSats, the four spectral bands were sufficient at generating satisfactory mapping products, as revealed by the accuracy analysis, which is an indication that the spectral and radiometric limitations were offset by the very high temporal and spatial resolutions.
Among the classification approaches evaluated, U-Net was the one that best used the spatial resolution of the PlanetScope data. Because it is a semantic segmentation approach, it was able to better populate the regions that were effectively pastures, in general producing a more spatially consistent mapping than the other methods. On the other hand, its use depended on segment samples for the totality of the input data, which usually required a greater effort of visual interpretation and generation compared to the point samples.
Specifically regarding the modifications we implemented in the U-Net, we believe that incorporating the temporal dimension with early-fusion and simultaneously analyzing two images contributed to a better identification of pasture pixels since the images used (i.e., November and March) contained complementary spectral data related to the seasonal and occupation dynamics of the study area. As the selection of these images made use of knowledge of the mapping region, for a scenario of application in large geographic regions, the input images could be automatically defined using statistical criteria (e.g., variation of NDVI values over a temporal window [
33], or all available images could be effectively used as input [
28]). However, the use of all images obtained in, for example, 12 months could increase the cost of segment sample generation as it would be necessary to consider their respective spatial variations during this period due to landscape dynamics and LULC changes [
60,
61].
In this scenario, the classification approach with the LSTM might be more appropriate because using a 3 × 3 pixel window and all available images, it considers both the spatial and temporal dimensions, such that its training samples were less affected by landscape dynamics and LULC changes. In this sense, our implementation allowed for the spatial window to be expanded to better consider the geographical context of the analyzed pixels. In fact, for this modification to improve the spatial consistency of the pasture classification results, our point sample set would have to be expanded beyond pure pixels (i.e., unmixed LULC class pixels).
Of all the evaluated approaches, the LSTM was the least dependent on some prior knowledge of the region and/or mapping classes, characteristics that make it very suitable for large-scale operational mappings. It is noteworthy that its classification efficiency has the potential to improve with a larger number of training samples, something that could be investigated by future studies. Because it is a per-pixel time-series approach, this classification model analyzed a data volume substantially larger than U-Net in the prediction step. Since, within the scope of satellite data classification, this step is embarrassingly parallel [
62], its execution time could be dramatically reduced through a high performance processing solution (e.g., in-house grid computing, Google Earth Engine).
5. Conclusions
The rapid growth of satellites orbiting the planet was responsible for generating a massive volume of data on the earth’s surface with the most varied scales and characteristics. At the same time, new deep-learning-based classification algorithms have been developed and are considered the state-of-the-art in many application domains. Given the possibility of using these algorithms on this growing volume of satellite data, this work evaluated three classification approaches based on PlanetScope images and the operational generation of mapping products for the pasture class. Our results indicated a better classification efficiency using deep learning approaches (i.e., LSTM and U-Net) over random forest, a conventional machine learning approach. However, additional comparison among these approaches are necessary, involving other studies areas and LULC classes. Nevertheless, considering the difficulty involved in the identification of pasture areas and the results obtained, we can infer that these classification approaches could be used to map other LULC classes (e.g., crops, planted forest, forest formations, water), taking into account new efforts to collect reference samples and training specialized models according to the classes of interest.
Likewise, and despite the fact that the generalization of these approaches to other geographic regions and/or mapping periods requires further investigations, the neural network architectures developed in this study could be used to map large geographic regions (e.g., the entire Brazilian territory), considering private (e.g., PlanetScope) and public (e.g., Sentinel 2, Landsat 8) satellite data. In this scenario, the strategy that we defined could be used to generate samples for the entire region and/or mapping period, allowing the training of classification models to use some level of temporal and/or geographic stratification, which is a procedure previously used to map pastures with random forest [
15,
33]. Finally, the effective rationalization of such classification approaches depends on high-performance processing solutions capable of combining GPU processing and access to a large volume of satellite data, which is likely to be available to governmental, private and research organizations in the next few years.