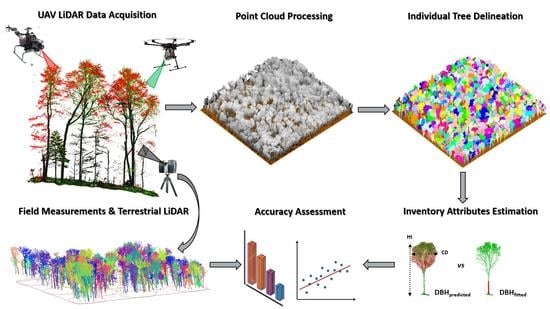
Estimation of Northern Hardwood Forest Inventory Attributes Using UAV Laser Scanning (ULS): Transferability of Laser Scanning Methods and Comparison of Automated Approaches at the Tree- and Stand-Level
Abstract
:1. Introduction
2. Materials
2.1. Study Site
2.2. Field Inventory
2.3. Terrestrial Laser Scanning (TLS) Data
2.4. UAV Laser Scanning (ULS) Data
2.5. Airborne Laser Scanning (ALS) Data
3. Methods
3.1. Experimental Design
- Uncertainties when matching field measurements (location and DBH in the field) with aerial 3D point clouds that were collected from above the canopy. The complex form of hardwood crowns leads to:
- difficult identifications of crown apices compared to coniferous trees (convoluted vs. conical crown shape);
- offsets from the base of the trunk for leaning and forked trees, which are quite common in hardwood stands;
- confused crown identifications with respect to their neighborhoods, since crowns are often interlocked.
3.2. Global Workflow
3.3. Data Co-Registration
3.4. Individual Tree Detection and Delineation (ITD)
3.4.1. Raster-Based ITD—SEGMA
3.4.2. Point Cloud-Based ITD—SimpleTree
3.5. Tree-Level Structural Attributes Estimation
3.5.1. Tree Height and Crown Diameter (CD)
- (i)
- vertically dividing the tree point cloud into 10-cm height clusters;
- (ii)
- fitting convex hull polygons to xy-coordinates of each cluster along the tree bole;
- (iii)
- calculating maximum Euclidean distance between the centroid of each convex hull and its vertices, and plotting the results along the z-axis (Figure 9A);
- (iv)
- identifying the CBH of the tree by fitting a segmented (piecewise) regression to the plotted points using the Segmented R package [107] (the CBH is defined as the lowest breakpoint (knot) of the segmented regression, which corresponds to the height where the regression slope starts to increase sharply because of the presence of branches) (Figure 9A “Breakpoint”); and
- (v)
- classifying the points above the CBH as belonging to the crown (Figure 9B), and using them to compute CD.
3.5.2. Diameter at Breast Height (DBH)
3.6. Stand-Level Inventory Attribute Estimation
3.7. Evaluation Methods
3.7.1. ITD Performance
3.7.2. Accuracy Assessment on Estimated Attributes
4. Results
4.1. ITD Performance
4.2. Tree-Level Structural Attribute Accuracy
4.3. Stand-Level Inventory Attribute Accuracy
4.4. Sensitivity Analysis
5. Discussion
5.1. Transferability of ITD Algorithms to ULS Data
5.2. Forest Inventory Attributes of an Uneven-Aged Hardwood Stand Using ULS Data
- Predicting DBH using height and CD allometry (Equation (1)) from raster-based ITD trees;
- Predicting DBH using height and CD allometry (Equation (1)) from point cloud-based ITD trees;
- Estimating DBH using cylinder-fitting algorithm.
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Airborne Laser Scanning |
| ALS-Raster | ALS dataset delineated using a Raster-based ITD |
| AGB | Above Ground Level |
| BA | Basal Area |
| CBH | Crown Based Height |
| CD | Crown Diameter |
| CHM | Canopy Height Model |
| DBH | Diameter at Breast Height |
| DBHfit | DBH estimated from cylinder fitting technique |
| DBHpred | DBH predicted from allometric models |
| DBHTLS | DBH derived from TLS data |
| DTM | Digital Terrain Model |
| FI | Forest Inventory |
| GCP | Ground Control Point |
| GNSS | Global Navigation Satellite System |
| Ht | Height |
| IMU | Inertial Measurement Unit |
| ITD | Individual Tree Detection and Delineation |
| LiDAR | Light Detection and Ranging |
| RMSE | Root Mean Square Error |
| TLS | Terrestrial Laser Scanning |
| UAV | Unmanned Aerial Vehicle |
| ULS | UAV Laser Scanning |
| ULS-R | ULS-Riegl Vux-1LR |
| ULS-R-Raster | ULS-R dataset delineated using a Raster-based ITD |
| ULS-V | ULS-Velodyne HDL-32E |
| ULS-V-Pcloud | ULS-V dataset delineated using a Point cloud-based ITD |
| ULS-V-Raster | ULS-V dataset delineated using a Raster-based ITD |
References
- Church, R.L.; Murray, A.T.; Barber, K.H. Forest Planning at the Tactical Level. Ann. Oper. Res. 2000, 95, 3–18. [Google Scholar] [CrossRef]
- Andersson, D. Approaches to Integrated Strategic/Tactical Forest Planning. 2005, pp. 1–29. Available online: https://pub.epsilon.slu.se/928/ (accessed on 12 February 2019).
- Luoma, V.; Saarinen, N.; Wulder, M.A.; White, J.C.; Vastaranta, M.; Holopainen, M.; Hyyppä, J. Assessing Precision in Conventional Field Measurements of Individual Tree Attributes. Forests 2017, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Brang, P.; Spathelf, P.; Larsen, J.B.; Bauhus, J.; Bončína, A.; Chauvin, C.; Drössler, L.; García-Güemes, C.; Heiri, C.; Kerr, G.; et al. Suitability of Close-to-Nature Silviculture for Adapting Temperate European Forests to Climate Change. Forestry 2014, 87, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Diaci, J.; Kerr, G.; O’Hara, K. Twenty-First Century Forestry: Integrating Ecologically Based, Uneven-Aged Silviculture with Increased Demands on Forests. Forestry 2011, 84, 463–465. [Google Scholar] [CrossRef] [Green Version]
- Banaś, J.; Ziȩba, S.; Bujoczek, L. An Example of Uneven-Aged Forest Management for Sustainable Timber Harvesting. Sustainability 2018, 10, 3305. [Google Scholar] [CrossRef] [Green Version]
- Nolet, P.; Kneeshaw, D.; Messier, C.; Béland, M. Comparing the Effects of Even- and Uneven-Aged Silviculture on Ecological Diversity and Processes: A Review. Ecol. Evol. 2018, 8, 1217–1226. [Google Scholar] [CrossRef]
- Leak, W.B.; Yamasaki, M.; Holleran, R. Silvicultural Guide for Northern Hardwoods in the Northeast. General Technical Report NRS-132; United States Department of Agriculture: Delaware, OH, USA, 2014; p. 46. Available online: https://www.nrs.fs.fed.us/pubs/45874 (accessed on 20 June 2020).
- Næsset, E. Practical Large-Scale Forest Stand Inventory Using a Small-Footprint Airborne Scanning Laser. Scand. J. For. Res. 2004, 19, 164–179. [Google Scholar] [CrossRef]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.P. Generalizing Predictive Models of Forest Inventory Attributes Using an Area-Based Approach with Airborne LiDAR Data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- White, J.C.; Tompalski, P.; Vastaranta, M.; Wulder, M.A.; Saarinen, N.; Stepper, C.; Coops, N.C. A Model Development and Application Guide for Generating an Enhanced Forest Inventory Using Airborne Laser Scanning Data and an Area-Based Approach. 2017. Available online: https://cfs.nrcan.gc.ca/publications?id=38945 (accessed on 10 January 2019).
- Woods, M.; Pitt, D.; Penner, M.; Lim, K.; Nesbitt, D.; Etheridge, D.; Treitz, P. Operational Implementation of a LiDAR Inventory in Boreal Ontario. For. Chron. 2011, 87, 512–528. [Google Scholar] [CrossRef]
- Treitz, P.; Lim, K.; Woods, M.; Pitt, D.; Nesbitt, D.; Etheridge, D. LiDAR Sampling Density for Forest Resource Inventories in Ontario, Canada. Remote Sens. 2012, 4, 830–848. [Google Scholar] [CrossRef] [Green Version]
- Brandtberg, T.; Warner, T.A.; Landenberger, R.E.; McGraw, J.B. Detection and Analysis of Individual Leaf-off Tree Crowns in Small Footprint, High Sampling Density Lidar Data from the Eastern Deciduous Forest in North America. Remote Sens. Environ. 2003, 85, 290–303. [Google Scholar] [CrossRef]
- Hyyppä, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A Segmentation-Based Method to Retrieve Stem Volume Estimates from 3-D Tree Height Models Produced by Laser Scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Nelson, R.F. Measuring Individual Tree Crown Diameter with Lidar and Assessing Its Influence on Estimating Forest Volume and Biomass. Can. J. Remote Sens. 2003, 29, 564–577. [Google Scholar] [CrossRef]
- Popescu, S.C. Estimating Biomass of Individual Pine Trees Using Airborne Lidar. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree Species Classification and Estimation of Stem Volume and DBH Based on Single Tree Extraction by Exploiting Airborne Full-Waveform LiDAR Data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Gleason, C.J.; Im, J. Forest Biomass Estimation from Airborne LiDAR Data Using Machine Learning Approaches. Remote Sens. Environ. 2012, 125, 80–91. [Google Scholar] [CrossRef]
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L.; Tomé, M.; Soares, P. 3-D Mapping of a Multi-Layered Mediterranean Forest Using ALS Data. Remote Sens. Environ. 2012, 121, 210–223. [Google Scholar] [CrossRef]
- Coomes, D.A.; Dalponte, M.; Jucker, T.; Asner, G.P.; Banin, L.F.; Burslem, D.F.R.P.; Lewis, S.L.; Nilus, R.; Phillips, O.L.; Phua, M.H.; et al. Area-Based vs Tree-Centric Approaches to Mapping Forest Carbon in Southeast Asian Forests from Airborne Laser Scanning Data. Remote Sens. Environ. 2017, 194, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Guo, Q.; Li, W.; Flanagan, J. A Bottom-up Approach to Segment Individual Deciduous Trees Using Leaf-off Lidar Point Cloud Data. ISPRS J. Photogramm. Remote Sens. 2014, 94, 1–12. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in Automatic Individual Tree Crown Detection and Delineation-Evolution of LiDAR Data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef] [Green Version]
- Lindberg, E.; Holmgren, J. Individual Tree Crown Methods for 3D Data from Remote Sensing. Curr. For. Rep. 2017, 3, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Maltamo, M.; Næsset, E.; Vauhkonen, J. Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies. Manag. Ecosyst. 2014, 27, 2014. [Google Scholar]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating Individual Trees in a Savanna Woodland Using Small Footprint Lidar Data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Koch, B.; Heyder, U.; Weinacker, H. Detection of Individual Tree Crowns in Airborne LIDAR Data. Photogramm. Eng. Remote Sens. 2006, 72, 357–363. [Google Scholar] [CrossRef] [Green Version]
- Jucker, T.; Caspersen, J.; Chave, J.; Antin, C.; Barbier, N.; Bongers, F.; Dalponte, M.; van Ewijk, K.Y.; Forrester, D.I.; Haeni, M.; et al. Allometric Equations for Integrating Remote Sensing Imagery into Forest Monitoring Programmes. Glob. Chang. Biol. 2017, 23, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A New Method for Segmenting Individual Trees from the Lidar Point Cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Aubry-Kientz, M.; Dutrieux, R.; Ferraz, A.; Saatchi, S.; Hamraz, H.; Williams, J.; Coomes, D.; Piboule, A.; Vincent, G. A Comparative Assessment of the Performance of Individual Tree Crowns Delineation Algorithms from ALS Data in Tropical Forests. Remote Sens. 2019, 11, 1086. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Xu, S.; Elberink, S.O.; Vosselman, G. Individual Tree Crown Modeling and Change Detection from Airborne Lidar Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3467–3477. [Google Scholar] [CrossRef]
- Vega, C.; Hamrouni, A.; El Mokhtari, A.; Morel, M.; Bock, J.; Renaud, J.P.; Bouvier, M.; Durrieue, S. PTrees: A Point-Based Approach to Forest Tree Extractionfrom Lidar Data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 98–108. [Google Scholar] [CrossRef]
- Wang, Y.; Weinacker, H.; Koch, B. A Lidar Point Cloud Based Procedure for Vertical Canopy Structure Analysis and 3D Single Tree Modelling in Forest. Sensors 2008, 8, 3938–3951. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Ye, N.; Xu, S.; Zhu, F. A Supervoxel Approach to the Segmentation of Individual Trees from LiDAR Point Clouds. Remote Sens. Lett. 2018, 9, 515–523. [Google Scholar] [CrossRef]
- Raumonen, P.; Casella, E.; Calders, K.; Murphy, S.; Åkerblom, M.; Kaasalainen, M. Massive-Scale Tree Modelling from TLS Data. ISPRS Ann. Photogramm. Remote Sens. Spatial Inf. Sci. 2015, 2, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, J.; Spiecker, H.; Calders, K.; Disney, M.; Raumonen, P. SimpleTree —An Efficient Open Source Tool to Build Tree Models from TLS Clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Ravaglia, J.; Bac, A.; Fournier, R.A. Extraction of Tubular Shapes from Dense Point Clouds and Application to Tree Reconstruction from Laser Scanned Data. Comput. Graph. 2017, 66, 23–33. [Google Scholar] [CrossRef]
- Othmani, A.; Piboule, A.; Krebs, M.; Stolz, C. Towards Automated and Operational Forest Inventories with T-Lidar. In Proceedings of the 11th International Conference on LiDAR Applications for Assessing Forest Ecosystems (SilviLaser 2011), Hobart, Australia, 16–20 October 2011; Available online: https://hal.archives-ouvertes.fr/hal-00646403/ (accessed on 15 September 2018).
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International Benchmarking of Terrestrial Laser Scanning Approaches for Forest Inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Vastaranta, M.; Holopainen, M.; Yu, X.; Hyyppä, J.; Mäkinen, A.; Rasinmäki, J.; Melkas, T.; Kaartinen, H.; Hyyppä, H. Effects of Individual Tree Detection Error Sources on Forest Management Planning Calculations. Remote Sens. 2011, 3, 1614–1626. [Google Scholar] [CrossRef] [Green Version]
- Kaartinen, H.; Hyyppä, J.; Yu, X.; Vastaranta, M.; Hyyppä, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F.; et al. An International Comparison of Individual Tree Detection and Extraction Using Airborne Laser Scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef] [Green Version]
- Eysn, L.; Hollaus, M.; Lindberg, E.; Berger, F.; Monnet, J.M.; Dalponte, M.; Kobal, M.; Pellegrini, M.; Lingua, E.; Mongus, D.; et al. A Benchmark of Lidar-Based Single Tree Detection Methods Using Heterogeneous Forest Data from the Alpine Space. Forests 2015, 6, 1721–1747. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hyyppa, J.; Liang, X.; Kaartinen, H.; Yu, X.; Lindberg, E.; Holmgren, J.; Qin, Y.; Mallet, C.; Ferraz, A.; et al. International Benchmarking of the Individual Tree Detection Methods for Modeling 3-D Canopy Structure for Silviculture and Forest Ecology Using Airborne Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2016, 54, 5011–5027. [Google Scholar] [CrossRef] [Green Version]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, F.M.; Demol, M.; Disney, M.; et al. Terrestrial Laser Scanning in Forest Ecology: Expanding the Horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- Weinacker, H.; Koch, B.; Heyder, U. Development of Filtering, Segmentation and Modelling Modules for Lidar and Multispectral Data as a Fundament of an Automatic Forest Inventory System. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 36 Pt 8, W2. [Google Scholar]
- Jing, L.; Hu, B.; Li, J.; Noland, T. Automated Delineation of Individual Tree Crowns from Lidar Data by Multi-Scale Analysis and Segmentation. Photogramm. Eng. Remote Sens. 2012, 78, 1275–1284. [Google Scholar] [CrossRef]
- Smreček, R.; Sačkov, I.; Michňová, Z.; Tuček, J. Automated Tree Detection and Crown Delineation Using Airborne Laser Scanner Data in Heterogeneous East-Central Europe Forest with Different Species Mix. J. For. Res. 2017, 28, 1049–1059. [Google Scholar] [CrossRef]
- Shendryk, I.; Broich, M.; Tulbure, M.G.; Alexandrov, S.V. Bottom-up Delineation of Individual Trees from Full-Waveform Airborne Laser Scans in a Structurally Complex Eucalypt Forest. Remote Sens. Environ. 2016, 173, 69–83. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Watson, C.S. Evaluating Tree Detection and Segmentation Routines on Very High Resolution UAV LiDAR Ata. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7619–7628. [Google Scholar] [CrossRef]
- Davison, S.; Donoghue, D.N.M.; Galiatsatos, N. The Effect of Leaf-on and Leaf-off Forest Canopy Conditions on LiDAR Derived Estimations of Forest Structural Diversity. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102160. [Google Scholar] [CrossRef]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Effects of Different Sensors and Leaf-on and Leaf-off Canopy Conditions on Echo Distributions and Individual Tree Properties Derived from Airborne Laser Scanning. Remote Sens. Environ. 2010, 114, 1445–1461. [Google Scholar] [CrossRef]
- Wasser, L.; Day, R.; Chasmer, L.; Taylor, A. Influence of Vegetation Structure on Lidar-Derived Canopy Height and Fractional Cover in Forested Riparian Buffers During Leaf-Off and Leaf-On Conditions. PLoS ONE 2013, 8, e54776. [Google Scholar] [CrossRef] [Green Version]
- Colomina, I.; Molina, P. Unmanned Aerial Systems for Photogrammetry and Remote Sensing: A Review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef] [Green Version]
- Torresan, C.; Berton, A.; Carotenuto, F.; Di Gennaro, S.F.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; Wallace, L. Forestry Applications of UAVs in Europe: A Review. Int. J. Remote Sens. 2017, 38, 2427–2447. [Google Scholar] [CrossRef]
- Guimarães, N.; Pádua, L.; Marques, P.; Silva, N.; Peres, E.; Sousa, J.J. Forestry Remote Sensing from Unmanned Aerial Vehicles: A Review Focusing on the Data, Processing and Potentialities. Remote Sens. 2020, 12, 1046. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Wang, Y.; Pyörälä, J.; Lehtomäki, M.; Yu, X.; Kaartinen, H.; Kukko, A.; Honkavaara, E.; Issaoui, A.E.I.; Nevalainen, O.; et al. Forest in Situ Observations Using Unmanned Aerial Vehicle as an Alternative of Terrestrial Measurements. For. Ecosyst. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Wallace, L.; Lucieer, A.; Watson, C.; Turner, D. Development of a UAV-LiDAR System with Application to Forest Inventory. Remote Sens. 2012, 4, 1519–1543. [Google Scholar] [CrossRef] [Green Version]
- Torresan, C.; Carotenuto, F.; Chiavetta, U.; Miglietta, F.; Zaldei, A.; Gioli, B. Individual Tree Crown Segmentation in Two-Layered Dense Mixed Forests from UAV Lidar Data. Drones 2020, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Gottfried, M.; Hollaus, M.; Glira, P.; Wieser, M.; Milenkovi, M.; Riegl, U.; Pfennigbauer, M. First Examples from the RIEGL VUX-SYS for Forestry Applications. Proc. SilviLaser 2015, 2015, 105–107. [Google Scholar]
- Brede, B.; Lau, A.; Bartholomeus, H.M.; Kooistra, L. Comparing RIEGL RiCOPTER UAV LiDAR Derived Canopy Height and DBH with Terrestrial LiDAR. Sensors 2017, 17, 2371. [Google Scholar] [CrossRef]
- Brede, B.; Calders, K.; Lau, A.; Raumonen, P.; Bartholomeus, H.M.; Herold, M.; Kooistra, L. Non-Destructive Tree Volume Estimation through Quantitative Structure Modelling: Comparing UAV Laser Scanning with Terrestrial LIDAR. Remote Sens. Environ. 2019, 233. [Google Scholar] [CrossRef]
- Belmonte, A.; Sankey, T.; Biederman, J.A.; Bradford, J.; Goetz, S.J.; Kolb, T.; Woolley, T. UAV-Derived Estimates of Forest Structure to Inform Ponderosa Pine Forest Restoration. Remote Sens. Ecol. Conserv. 2020, 6, 181–197. [Google Scholar] [CrossRef]
- Ravanel, L.; Bodin, X.; Deline, P. Using Terrestrial Laser Scanning for the Recognition and Promotion of High-Alpine Geomorphosites. Geoheritage 2014, 6, 129–140. [Google Scholar] [CrossRef]
- Jaakkola, A.; Hyyppä, J.; Kukko, A.; Yu, X.; Kaartinen, H.; Lehtomäki, M.; Lin, Y. A Low-Cost Multi-Sensoral Mobile Mapping System and Its Feasibility for Tree Measurements. ISPRS J. Photogramm. Remote Sens. 2010, 65, 514–522. [Google Scholar] [CrossRef]
- Kucharczyk, M.; Hugenholtz, C.H.; Zou, X. UAV-LiDAR Accuracy in Vegetated Terrain. J. Unmanned Veh. Syst. 2018, 6, 212–234. [Google Scholar] [CrossRef]
- Torresan, C.; Berton, A.; Carotenuto, F.; Chiavetta, U.; Miglietta, F.; Zaldei, A.; Gioli, B. Development and Performance Assessment of a Low-Cost UAV Laser Scanner System (LasUAV). Remote Sens. 2018, 10, 1094. [Google Scholar] [CrossRef] [Green Version]
- Sofonia, J.J.; Phinn, S.; Roelfsema, C.; Kendoul, F.; Rist, Y. Modelling the Effects of Fundamental UAV Flight Parameters on LiDAR Point Clouds to Facilitate Objectives-Based Planning. ISPRS J. Photogramm. Remote Sens. 2019, 149, 105–118. [Google Scholar] [CrossRef]
- Vepakomma, U.; Cormier, D. Potential of Multi-Temporal UAV-Borne Lidar in Assessing Effectiveness of Silvicultural Treatments. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, 42, 393–397. [Google Scholar] [CrossRef] [Green Version]
- Balsi, M.; Esposito, S.; Fallavollita, P.; Nardinocchi, C. Single-Tree Detection in High-Density LiDAR Data from UAV-Based Survey. Eur. J. Remote Sens. 2018, 51, 679–692. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yang, B.; Cong, Y.; Cao, L.; Fu, X.; Dong, Z. 3D Forest Mapping Using a Low-Cost UAV Laser Scanning System: Investigation and Comparison. Remote Sens. 2019, 11, 717. [Google Scholar] [CrossRef] [Green Version]
- Bruggisser, M.; Hollaus, M.; Otepka, J.; Pfeifer, N. Influence of ULS Acquisition Characteristics on Tree Stem Parameter Estimation. ISPRS J. Photogramm. Remote Sens. 2020, 168, 28–40. [Google Scholar] [CrossRef]
- Wallace, L.; Musk, R.; Lucieer, A. An Assessment of the Repeatability of Automatic Forest Inventory Metrics Derived from UAV-Borne Laser Scanning Data. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7160–7169. [Google Scholar] [CrossRef]
- Jaakkola, A.; Hyyppä, J.; Yu, X.; Kukko, A.; Kaartinen, H.; Liang, X.; Hyyppä, H.; Wang, Y. Autonomous Collection of Forest Field Reference—The Outlook and a First Step with UAV Laser Scanning. Remote Sens. 2017, 9, 785. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating Forest Structural Attributes Using UAV-LiDAR Data in Ginkgo Plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Zaforemska, A.; Xiao, W.; Gaulton, R. Individual Tree Detection from UAV Lidar Data in a Mixed Species Woodland. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 657–663. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Pyörälä, J.; Liang, X.; Lehtomäki, M.; Kukko, A.; Yu, X.; Kaartinen, H.; Hyyppä, J. In Situ Biomass Estimation at Tree and Plot Levels: What Did Data Record and What Did Algorithms Derive from Terrestrial and Aerial Point Clouds in Boreal Forest. Remote Sens. Environ. 2019, 232, 111309. [Google Scholar] [CrossRef]
- Sačkov, I.; Santopuoli, G.; Bucha, T.; Lasserre, B.; Marchetti, M. Forest Inventory Attribute Prediction Using Lightweight Aerial Scanner Data in a Selected Type of Multilayered Deciduous Forest. Forests 2016, 7, 307. [Google Scholar] [CrossRef] [Green Version]
- Wieser, M.; Mandlburger, G.; Hollaus, M.; Otepka, J.; Glira, P.; Pfeifer, N. A Case Study of UAS Borne Laser Scanning for Measurement of Tree Stem Diameter. Remote Sens. 2017, 9, 1154. [Google Scholar] [CrossRef] [Green Version]
- Ecosystem Classification Working Group Our Landscape Heritage: The Story of Ecological Land Classification in New Brunswick. 2007, pp. 1–16. Available online: https://www2.gnb.ca/ (accessed on 20 June 2019).
- Todd, W. Bowersox, The Practice of Silviculture—Applied Forest Ecology, Ninth Edition. Forest Sci. 1997, 43, 455–456. [Google Scholar] [CrossRef]
- Alba, M.; Scaioni, M. Comparison of Techniques for Terrestrial Laser Scanning Data Georeferencing Applied to 3D Modeling of Cultural Heritage. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2007, 36, 25–33. [Google Scholar]
- GeoNB. Province of New Brunswick’s Gateway to Geographic Information. Available online: http://www.snb.ca/geonb1/e/index-E.asp (accessed on 5 January 2020).
- Larjavaara, M.; Muller-Landau, H.C. Measuring Tree Height: A Quantitative Comparison of Two Common Field Methods in a Moist Tropical Forest. Methods Ecol. Evol. 2013, 4, 793–801. [Google Scholar] [CrossRef]
- Wang, Y.; Lehtomäki, M.; Liang, X.; Pyörälä, J.; Kukko, A.; Jaakkola, A.; Liu, J.; Feng, Z.; Chen, R.; Hyyppä, J. Is Field-Measured Tree Height as Reliable as Believed—A Comparison Study of Tree Height Estimates from Field Measurement, Airborne Laser Scanning and Terrestrial Laser Scanning in a Boreal Forest. ISPRS J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Jurjević, L.; Liang, X.; Gašparović, M.; Balenović, I. Is Field-Measured Tree Height as Reliable as Believed—Part II, A Comparison Study of Tree Height Estimates from Conventional Field Measurement and Low-Cost Close-Range Remote Sensing in a Deciduous Forest. ISPRS J. Photogramm. Remote Sens. 2020, 169, 227–241. [Google Scholar] [CrossRef]
- Metz, J.Ô.; Seidel, D.; Schall, P.; Scheffer, D.; Schulze, E.D.; Ammer, C. Crown Modeling by Terrestrial Laser Scanning as an Approach to Assess the Effect of Aboveground Intra- and Interspecific Competition on Tree Growth. For. Ecol. Manag. 2013, 310, 275–288. [Google Scholar] [CrossRef]
- Seidel, D.; Fleck, S.; Leuschner, C. Analyzing Forest Canopies with Ground-Based Laser Scanning: A Comparison with Hemispherical Photography. Agric. For. Meteorol. 2012, 154–155, 1. [Google Scholar] [CrossRef]
- Srinivasan, S.; Popescu, S.C.; Eriksson, M.; Sheridan, R.D.; Ku, N.W. Terrestrial Laser Scanning as an Effective Tool to Retrieve Tree Level Height, Crown Width, and Stem Diameter. Remote Sens. 2015, 7, 1877–1896. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial Laser Scanning in Forest Inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Levick, S.R.; Whiteside, T.; Loewensteiner, D.A.; Rudge, M.; Bartolo, R. Leveraging Tls as a Calibration and Validation Tool for Mls and Uls Mapping of Savanna Structure and Biomass at Landscape-Scales. Remote Sens. 2021, 13, 257. [Google Scholar] [CrossRef]
- Rajendra, Y.D.; Mehrotra, S.C.; Kale, K.V.; Manza, R.R.; Dhumal, R.K.; Nagne, A.D.; Vibhute, A.D. Evaluation of Partially Overlapping 3D Point Cloud’s Registration by Using ICP Variant and Cloudcompare. Int. Arch. Photogramm. Remote Sens. Spatial Inf. Sci. 2014, 40, 891–897. [Google Scholar] [CrossRef] [Green Version]
- Hilker, T.; Coops, N.C.; Culvenor, D.S.; Newnham, G.; Wulder, M.A.; Bater, C.W.; Siggins, A. A Simple Technique for Co-Registration of Terrestrial LiDAR Observations for Forestry Applications. Remote Sens. Lett. 2012, 3, 239–247. [Google Scholar] [CrossRef]
- Theiler, P.W.; Wegner, J.D.; Schindler, K. Keypoint-Based 4-Points Congruent Sets—Automated Marker-Less Registration of Laser Scans. ISPRS J. Photogramm. Remote Sens. 2014, 96, 149–163. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Ene, L.; Gupta, S.; Heinzel, J.; Holmgren, J.; Pitkänen, J.; Solberg, S.; Wang, Y.; Weinacker, H.; Hauglin, K.M.; et al. Comparative Testing of Single-Tree Detection Algorithms under Different Types of Forest. Forestry 2012, 85, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Roussel, J.R.; Auty, D.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.H.; Meador, A.S.; Bourdon, J.F.; de Boissieu, F.; Achim, A. LidR: An R Package for Analysis of Airborne Laser Scanning (ALS) Data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- St-Onge, B.; Audet, F.-A.; Bégin, J. Characterizing the Height Structure and Composition of a Boreal Forest Using an Individual Tree Crown Approach Applied to Photogrammetric Point Clouds. Forests 2015, 6, 3899–3922. [Google Scholar] [CrossRef]
- Python Software Foundation. Python Language Reference, Version 2.7. Available online: http://www.python.org (accessed on 1 February 2019).
- Computree Core Team. Computree Platform. 2017. Available online: http://rdinnovation.onf.fr/computree (accessed on 20 May 2019).
- Stereńczak, K.; Będkowski, K.; Weinacker, H. Accuracy of Crown Segmentation and Estimation of Selected Trees and Forest Stand Parameters in Order to Resolution of Used DSM and NDSM Models Generated from Dense Small Footprint LIDAR Data. In Proceedings of the ISPRS Congress, Commission VI, WG VI/5, Beijing, China, 3–11 July 2008; pp. 27–32. [Google Scholar]
- Barnes, C.; Balzter, H.; Barrett, K.; Eddy, J.; Milner, S.; Suárez, J.C. Individual Tree Crown Delineation from Airborne Laser Scanning for Diseased Larch Forest Stands. Remote Sens. 2017, 9, 231. [Google Scholar] [CrossRef] [Green Version]
- Dijkstra, E. A Note on Two Problems in Connexion with Graphs. Numer. Math 1959, 1, 269–271. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, J.; Morhart, C.; Sheppard, J.; Spiecker, H.; Disney, M. Highly Accurate Tree Models Derived from Terrestrial Laser Scan Data: A Method Description. Forests 2014, 5, 1069–1105. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, J.; Wassenberg, M.; Spiecker, H.; Sun, D. Non Destructive Method for Biomass Prediction Combining TLS Derived Tree Volume and Wood Density. Forests 2015, 6, 1274–1300. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 5 January 2019).
- Schneider, R.; Calama, R.; Martin-Ducup, O. Understanding Tree-to-Tree Variations in Stone Pine (Pinus Pinea L.) Cone Production Using Terrestrial Laser Scanner. Remote Sens. 2020, 12, 173. [Google Scholar] [CrossRef] [Green Version]
- Muggeo, V.R.M. Segmented: An R Package to Fit Regression Models with Broken-Line Relationships. R. News 2008, 3, 343–344. [Google Scholar]
- Ravaglia, J.; Bac, A.; Piboule, A. Laser-Scanned Tree Stem Filtering for Forest Inventories Measurements. Digit. Herit. Int. Congr. 2013, 1, 649–652. [Google Scholar] [CrossRef] [Green Version]
- Koreň, M.; Hunčaga, M.; Chudá, J.; Mokroš, M.; Surový, P. The Influence of Cross-Section Thickness on Diameter at Breast Height Estimation from Point Cloud. ISPRS Int. J. Geo-Inf. 2020, 9, 495. [Google Scholar] [CrossRef]
- Pitkänen, J.; Maltamo, M. Adaptive Methods for Individual Tree Detection on Airborne Laser Based Canopy Height Model. Int. Arch. Photogramm. Remote Sens. Spatial Info. Sci. 2004, 36, 187–191. [Google Scholar]
- Tanhuanpää, T.; Saarinen, N.; Kankare, V.; Nurminen, K.; Vastaranta, M.; Honkavaara, E.; Karjalainen, M.; Yu, X.; Holopainen, M.; Hyyppä, J. Evaluating the Performance of High-Altitude Aerial Image-Based Digital Surface Models in Detecting Individual Tree Crowns in Mature Boreal Forests. Forests 2016, 7, 143. [Google Scholar] [CrossRef] [Green Version]
- Che, E.; Jung, J.; Olsen, M.J. Object Recognition, Segmentation, and Classification of Mobile Laser Scanning Point Clouds: A State of the Art Review. Sensors 2019, 19, 810. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Hu, X.; Chen, W.; Hong, Y.; Yang, M. Airborne LiDAR Remote Sensing for Individual Tree Forest Inventory Using Trunk Detection-Aided Mean Shift Clustering Techniques. Remote Sens. 2018, 10, 1078. [Google Scholar] [CrossRef] [Green Version]
- Dersch, S.; Heurich, M.; Krueger, N.; Krzystek, P. Combining Graph-Cut Clustering with Object-Based Stem Detection for Tree Segmentation in Highly Dense Airborne Lidar Point Clouds. ISPRS J. Photogramm. Remote Sens. 2021, 172, 207–222. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Luo, Z. Combining Trunk Detection with Canopy Segmentation to Delineate Single Deciduous Trees Using Airborne LiDAR Data. IEEE Access 2020, 8, 99783–99796. [Google Scholar] [CrossRef]
- Jaskierniak, D.; Lucieer, A.; Kuczera, G.; Turner, D.; Lane, P.N.J.; Benyon, R.G.; Haydon, S. Individual Tree Detection and Crown Delineation from Unmanned Aircraft System (UAS) LiDAR in Structurally Complex Mixed Species Eucalypt Forests. ISPRS J. Photogramm. Remote Sens. 2021, 171, 171–187. [Google Scholar] [CrossRef]
- Reitberger, J.; Schnörr, C.; Krzystek, P.; Stilla, U. 3D Segmentation of Single Trees Exploiting Full Waveform LIDAR Data. ISPRS J. Photogramm. Remote Sens. 2009, 64, 561–574. [Google Scholar] [CrossRef]
- Andersen, H.E.; Reutebuch, S.E.; McGaughey, R.J.; d’Oliveira, M.V.N.; Keller, M. Monitoring Selective Logging in Western Amazonia with Repeat Lidar Flights. Remote Sens. Environ. 2014, 151, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q. Modeling Aboveground Tree Woody Biomass Using National-Scale Allometric Methods and Airborne Lidar. ISPRS J. Photogramm. Remote Sens. 2015, 106, 95–106. [Google Scholar] [CrossRef]
- Budei, B.C.; St-Onge, B.; Hopkinson, C.; Audet, F.-A. Identifying the Genus or Species of Individual Trees Using a Three-Wavelength Airborne Lidar System. Remote Sens. Environ. 2017, 204, 632–647. [Google Scholar] [CrossRef]
- Degerickx, J.; Roberts, D.A.; McFadden, J.P.; Hermy, M.; Somers, B. Urban Tree Health Assessment Using Airborne Hyperspectral and LiDAR Imagery. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 26–38. [Google Scholar] [CrossRef] [Green Version]
- Olivier, M.D.; Robert, S.; Fournier, R.A. Response of Sugar Maple (Acer saccharum, Marsh.) Tree Crown Structure to Competition in Pure versus Mixed Stands. For. Ecol. Manag. 2016, 374, 20–32. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, K.; Zhu, Y.; Wang, X.; Yun, T. Individual Tree Crown Segmentation Directly from Uav-Borne Lidar Data Using the Pointnet of Deep Learning. Forests 2021, 12, 131. [Google Scholar] [CrossRef]
- Morsdorf, F.; Eck, C.; Zgraggen, C.; Imbach, B.; Schneider, F.D.; Kükenbrink, D. UAV-Based LiDAR Acquisition for the Derivation of High-Resolution Forest and Ground Information. Lead. Edge 2017, 36, 566–570. [Google Scholar] [CrossRef]
- Lindberg, E.; Eysn, L.; Hollaus, M.; Holmgren, J.; Pfeifer, N. Delineation of Tree Crowns and Tree Species Classification from Full-Waveform Airborne Laser Scanning Data Using 3-d Ellipsoidal Clustering. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3174–3181. [Google Scholar] [CrossRef] [Green Version]
- Ferraz, A.; Saatchi, S.; Mallet, C.; Meyer, V. Lidar Detection of Individual Tree Size in Tropical Forests. Remote Sens. Environ. 2016, 183, 318–333. [Google Scholar] [CrossRef]
- Hamraz, H.; Contreras, M.A.; Zhang, J. A Robust Approach for Tree Segmentation in Deciduous Forests Using Small-Footprint Airborne LiDAR Data. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 532–541. [Google Scholar] [CrossRef] [Green Version]
- Seidel, D.; Annighöfer, P.; Thielman, A.; Seifert, Q.E.; Thauer, J.H.; Glatthorn, J.; Ehbrecht, M.; Kneib, T.; Ammer, C. Predicting Tree Species From 3D Laser Scanning Point Clouds Using Deep Learning. Front. Plant Sci. 2021, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dalponte, M.; Frizzera, L.; Ørka, H.O.; Gobakken, T.; Næsset, E.; Gianelle, D. Predicting Stem Diameters and Aboveground Biomass of Individual Trees Using Remote Sensing Data. Ecol. Indic. 2018, 85, 367–376. [Google Scholar] [CrossRef]
- Duncanson, L.I.; Cook, B.D.; Hurtt, G.C.; Dubayah, R.O. An Efficient, Multi-Layered Crown Delineation Algorithm for Mapping Individual Tree Structure across Multiple Ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Lindberg, E.; Holmgren, J.; Olofsson, K.; Wallerman, J.; Olsson, H. Estimation of Tree Lists from Airborne Laser Scanning by Combining Single-Tree and Area-Based Methods. Int. J. Remote Sens. 2010, 31, 1175–1192. [Google Scholar] [CrossRef] [Green Version]
- Vauhkonen, J.; Mehtätalo, L. Matching Remotely Sensed and Field Measured Tree Size Distributions. Can. J. For. Res. 2014, 45, 353–363. [Google Scholar] [CrossRef]
- Ferraz, A.; Mallet, C.; Jacquemoud, S.; Goncalves, G.R.; Tome, M.; Soares, P.; Pereira, L.G.; Bretar, F. Canopy Density Model: A New ALS-Derived Product to Generate Multilayer Crown Cover Maps. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6776–6790. [Google Scholar] [CrossRef]
- Du, S.; Lindenbergh, R.; Ledoux, H.; Stoter, J.; Nan, L. AdTree: Accurate, Detailed, and Automatic Modelling of Laser-Scanned Trees. Remote Sens. 2019, 11, 2074. [Google Scholar] [CrossRef] [Green Version]
















| Parameter | ALS | ULS-R | ULS-V | TLS |
|---|---|---|---|---|
| Platform |  |  |  |  |
| Sensor | Riegl LMS Q680i | Riegl Vux-1LR | Velodyne HDL-32E | FARO Focus 3D S 120 |
| Acquisition conditions | Leaf-on (June 2017) | Leaf-on (August 2016) | Leaf-off (December 2015) | Leaf-off (May 2017) |
| Average flying altitude | 1100 m | 185 m | 40 m | na |
| Pulse Repetition Frequency | 310 kHz | 600 kHz | 700 kHz | 244 kHz |
| Beam divergence | 0.5 mrad | 0.5 mrad | 3 mrad | 0.19 mrad |
| Field of view | [+30° to −30°] | [+40° to −40°] | [+ 10° to −30°] V X360° H | 300° V X360° H |
| Accuracy | 0.28 m @ 1100 m | 1.5 cm @ 50 m | 2.5 cm @ 50 m | 6.3 mm @ 10 m |
| Wavelength | 1550 nm | 1550 nm | 903 nm | 905 nm |
| Echoes/pulse | 5 | 7 | 2 | 1 |
| Point density | 27 points/m2 | 353 points/m2 | 1585 points/m2 | 60 K points/m2 |
| Criterion | Height Test & Distance Test | Distance Test |
|---|---|---|
| 1 | < 10 m & ∆H < 2.5 m | < 3 m |
| 2 | 10 m ≤ < 15 m & ∆H < 3 m | < 3.5 m |
| 3 | ≥ 15 m & ∆H < 4 m | < 4 m |
| Dataset | Canopy Condition | ITD Approach | DBH Approach | |||
|---|---|---|---|---|---|---|
| Leaf-On | Leaf-Off | SEGMA | SimpleTree | Predicted | Fitted | |
| ALS-Raster | x | x | x | |||
| ULS-R-Raster | x | x | x | |||
| ULS-V-Raster | x | x | x | |||
| ULS-V-Pcloud | x | x | x | |||
| FI & TLS (Ref) | x | x | x | |||
| Dataset | Acquisition Parameters | ITD Approach | by Height Class | ||||
|---|---|---|---|---|---|---|---|
| [6–12[ m | [12–18[ m | ≥18 m | Total | ||||
| ALS-Raster | Leaf-on (27 pts/m2) | SEGMA | 275 (58%) | 2 (5%) | 18 (15%) | 88 (91%) | 108 (42%) |
| ULS-R-Raster | Leaf-on (353 pts/m2) | SEGMA | 273 (57%) | 1 (2%) | 15 (13%) | 87 (90%) | 103 (40%) |
| ULS-V-Raster | Leaf-off (1585 pts/m2) | SEGMA | 346 (73%) | 4 (9%) | 28 (24%) | 96 (99%) | 128 (50%) |
| ULS-V-PCloud | SimpleTree | 340 (71%) | 22 (51%) | 70 (59%) | 91 (94%) | 183 (71%) | |
| FI & TLS (ref) | Leaf-off (60 k pts/m2) | SimpleTree & Manual ITD | 477 | 43 | 118 | 97 | 258 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vandendaele, B.; Fournier, R.A.; Vepakomma, U.; Pelletier, G.; Lejeune, P.; Martin-Ducup, O. Estimation of Northern Hardwood Forest Inventory Attributes Using UAV Laser Scanning (ULS): Transferability of Laser Scanning Methods and Comparison of Automated Approaches at the Tree- and Stand-Level. Remote Sens. 2021, 13, 2796. https://doi.org/10.3390/rs13142796
Vandendaele B, Fournier RA, Vepakomma U, Pelletier G, Lejeune P, Martin-Ducup O. Estimation of Northern Hardwood Forest Inventory Attributes Using UAV Laser Scanning (ULS): Transferability of Laser Scanning Methods and Comparison of Automated Approaches at the Tree- and Stand-Level. Remote Sensing. 2021; 13(14):2796. https://doi.org/10.3390/rs13142796
Chicago/Turabian StyleVandendaele, Bastien, Richard A. Fournier, Udayalakshmi Vepakomma, Gaetan Pelletier, Philippe Lejeune, and Olivier Martin-Ducup. 2021. "Estimation of Northern Hardwood Forest Inventory Attributes Using UAV Laser Scanning (ULS): Transferability of Laser Scanning Methods and Comparison of Automated Approaches at the Tree- and Stand-Level" Remote Sensing 13, no. 14: 2796. https://doi.org/10.3390/rs13142796
APA StyleVandendaele, B., Fournier, R. A., Vepakomma, U., Pelletier, G., Lejeune, P., & Martin-Ducup, O. (2021). Estimation of Northern Hardwood Forest Inventory Attributes Using UAV Laser Scanning (ULS): Transferability of Laser Scanning Methods and Comparison of Automated Approaches at the Tree- and Stand-Level. Remote Sensing, 13(14), 2796. https://doi.org/10.3390/rs13142796