1. Introduction
Hyperspectral images (HSI) contain hundreds of continuous and diverse bands rich in spectral and spatial information, which can distinguish land-cover types more efficiently compared with ordinary remote sensing images [
1,
2]. In recent years, Hyperspectral images classification (HSIC) has become one of the most important tasks in the field of remote sensing with wide application in scenarios such as urban planning, geological exploration, and agricultural monitoring [
3,
4,
5,
6].
Originally, models such as support vector machines (SVM) [
7], logistic regression (LR) [
8] and and k-nearest neighbors algorithm (KNN) [
9], have been widely used in HSI classification tasks for their intuitive outcomes. However, most of them only utilize handcrafted features, which fail to embody the distribution characteristics of different objects. To solve this problem, a series of deep discriminative models, such as convolutional neural networks (CNNs) [
10,
11,
12], recurrent neural network (RNN) [
13] and Deep Neural Networks (DNN) [
14] have been proposed to optimize the classification results by fully utilizing and abstracting the limited data. Though having gained great progress, these methods only analyze the spectral characteristics through an end-to-end neural network without full consideration of special properties contained in HSI. Therefore, the extraction of high-level and abstract features in HSIC remains a challenging task. Meanwhile, the jointed spectral-spatial features extraction methods [
15,
16] have aroused wide interest in Geosciences and Remote Sensing community [
17]. Du proposed a jointed network to extract spectral and spatial features with dimensionality reduction [
18]. Zhao et al. proposed a hybrid spectral CNN (HybridSN) to better extract double-way features [
19], which combined spectral-spatial 3D-CNN with spatial 2D-CNN to improve the classification accuracy.
Although the methods above enhance the abilities of spectral and spatial features extraction, they are still based on the discriminative model in essence, which can neither calculate prior probability nor describe the unique features of HSI data. In addition, the access to acquire HSI data is very expensive and scarce, requiring huge human resources to label the samples by field investigation. These characteristics make it impractical to obtain enough markable samples for training. Therefore, the deep generative models have emerged at the call of the time. Variational auto encoder (VAE) [
20] and generative adversarial network (GAN) [
21] are the representative methods of generative models.
Liu [
22] and Su [
23] used VAEs to ensure the diversity of the generated data that were sampled from the latent space. However, the generated HSI virtual samples are often ambiguous, which cannot guarantee similarities with the real HSI data. Therefore, GANs have also been applied for HSI generation to improve the quality of generated virtual data. GANs strengthen the ability of discriminators to distinguish the true data sources from the false by introducing “Nash equilibrium” [
24,
25,
26,
27,
28,
29]. For example, Zhan [
30] designed a 1-D GAN (HSGAN) to generate the virtual HSI pixels similar to the real ones, thus improving the performance of the classifier. Feng [
31] devised two generators to generate 2D-spatial and 1D-spectral information respectively. Zhu [
32] exploited 1D-GAN and 3D-GAN architectures to enhance the classification performance. However, GANs are prone to mode collapse, resulting in poor generalization ability of HSI classification.
To overcome the limitations of VAEs and GANs, VAE-GAN jointed framework has been proposed for HSIC. Wang proposed a conditional variational autoencoder with an adversarial training process for HSIC (CVA
E) [
33]. In this work, GAN was spliced with VAE to realize high-quality restoration of the samples and achieve diversity. Tao et al. [
34] proposed the semi-supervised variational generative adversarial networks with a collaborative relationship between the generation network and the classification network to produce meaningful samples that contribute to the final classification. To sum up, in VAE-GAN frameworks, VAE focuses on encoding the latent space, providing creativity of generated samples, while GAN concentrates on replicating the data, contributing to the high quality of virtual samples.
Spectral and spatial are two typical characteristics of HSI, both of which must be taken into account for HSIC. Nevertheless, the distributions of spectral and spatial features are not identical. Therefore, it is difficult to cope with such a complex situation for a single encoder in VAEs. Meanwhile, most of the existing generative methods use spectral and spatial features respectively for HSIC, which affects the generative model to generate realistic virtual samples. In fact, the spectral and spatial features are closely correlated, which cannot be treated separately. Interaction between spectral and spatial information should be established to refine the generated virtual samples for better classification performance.
In this paper, a variational generative adversarial network with crossed spatial and spectral interactions (CSSVGAN) was proposed for HSIC, which consists of a dual-branch variational Encoder, a crossed interactive Generator, and a Discriminator stuck together with a classifier. The dual-branch variational Encoder maps spectral and spatial information to different latent spaces. The crossed interactive Generator reconstructs the spatial and spectral samples from the latent spectral and spatial distribution in a crossed manner. Notably, the intersectional generation process promotes the consistency of learned spatial and spectral features and simulates the highly correlated spatial and spectral characteristics of true HSI. The Discriminator receives the samples from both generator and original training data to distinguish the authenticity of the data. To sum up, the variational Encoder ensures diversity, and the Generator guarantees authenticity. The two components place higher demands on the Discriminator to achieve better classification performance.
Compared with the existing literature, this paper is expected to make the following contributions:
The dual-branch variational Encoder in the jointed VAE-GAN framework is developed to map spectral and spatial information into different latent spaces, provides discriminative spectral and spatial features, and ensures the diversity of generated virtual samples.
The crossed interactive Generator is proposed to improve the quality of generated virtual samples, which exploits the consistency of learned spatial and spectral features to imitate the highly correlated spatial and spectral characteristics of HSI.
The variational generative adversarial network with crossed spatial and spectral interactions is proposed for HSIC, where the diversity and authenticity of generated samples are enhanced simultaneously.
Experimental results on the three public datasets demonstrate that the proposed CSSVGAN achieves better performance compared with other well-known models.
The remainder of this paper is arranged as follows.
Section 2 introduces VAEs and GANs.
Section 3 provides the details of the CSSVGAN framework and the crossed interactive module.
Section 4 evaluates the performance of the proposed CSSVGAN through comparison with other methods. The results of the experiment are discussed in
Section 5 and the conclusion is given in
Section 6.
3. Methodology
3.1. The Overall Framework of CSSVGAN
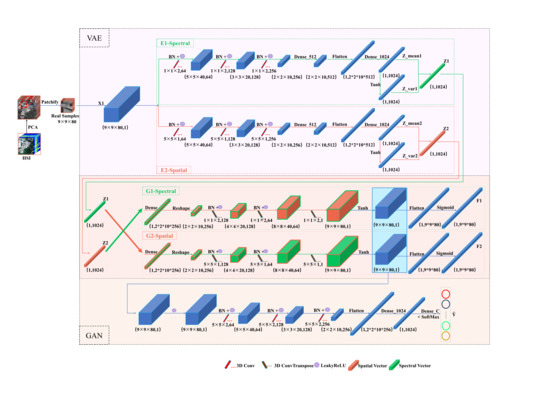
The overall framework of CSSVGAN is shown in
Figure 3. In the process of data preprocessing, assuming that HSI cuboid
X contains
n pixels; the spectral band of each pixel is defined as
; and
X can be expressed as
. Then HSI is divided into several patch cubes of the same size. The labeled pixels are marked as
, and the unlabeled pixels are marked as
. Among them,
s,
and
stand for the adjacent spatial sizes of HSI cuboids, the number of labeled samples and the number of unlabeled samples respectively, and
n equals to
plus
.
It is noteworthy that HSI classification is developed at the pixel level. Therefore, in this paper, the CSSVGAN framework uses a cube composed of patches of size as the inputs of the Encoder, where p denotes the spectral bands of each pixel. Then a tensor represents the variables and outputs of each layer. Firstly, the spectral latent variable and the spatial latent variable are obtained by taking the above as input into the dual-branch variational Encoder. Secondly, these two inputs are taken to the crossed interactive Generator module to obtain the virtual data and . Finally, the data are mixed with into the Discriminator for adversarial training to get the predicted classification results by the classifier.
3.2. The Dual-Branch Variational Encoder in CSSVGAN
In the CSSVGAN model mentioned above, the Encoder (
Figure 4) is composed of a dual-branch spatial feature extraction
and a spectral feature extraction
to generate more diverse samples. In the
module, the size of the 3D convolution kernel is
, the stride is
and the spectral features are marked as
. The implementation details are described in
Table 1. Identically, in the
module, the 3D convolution kernels, the strides and the spatial features are presented by
,
and
respectively, as described in
Table 2.
Meanwhile, to ensure the consistent distribution of samples and original data, KL divergence principle is utilized to constrain
and
separately. Assuming that the mean and variance of
are expressed as
and
, the loss function in the training process is as follows:
where
is the posterior distribution of potential eigenvectors in the Encoder module, and its calculation is based on the Bayesian formula as shown below. But when the dimension of
Z is too high, the calculation of
is not feasible. At this time, a known distribution
is required to approximate
, which is given by KL divergence. By minimizing KL divergence, the approximate
can be obtained.
and
represent the parameters of distribution function
p and
q separately.
Formula (6) in the back is provided with a constant term , the entropy of empirical distribution . The advantage of it is that the optimization objective function is more explicit, that is, when is equal to , KL dispersion can be minimized.
3.3. The Crossed Interactive Generator in CSSVGAN
In CSSVGAN, the crossed interactive Generator module plays a role in data restoration of VAE and data expansion of GAN, which includes the spectral Generator and the spatial Generator in the crossed manner. accepts the spatial latent variables to generate spectral virtual data , and accepts the spectral latent variables to generate spatial virtual data .
As shown in
Figure 5, the 3D convolution of spectral Generator
is
that uses
strides to convert the spatial latent variables
to the generated samples. Similarly, the spatial Generator
with
convolution uses
strides to transform the spectral latent variables
into generated samples. Therefore, the correlation between spectral and spatial features in HSI can be fully considered to further improve the quality and authenticity of the generated samples. The implementation details of
and
are described in
Table 3 and
Table 4.
Because the mechanism of GAN is that the Generator and Discriminator are against each other before reaching the Nash equilibrium, the Generator has two target functions, as shown below.
where
n is the number of samples,
,
means the label of virtual samples, and
represents the label of the original data corresponding to
. The above formula makes the virtual samples generated by crossed interactive Generator as similar as possible to the original data.
is a logarithmic loss function and can be applied to the binary classification task. Where y is the label (either true or false), and is the probability that N sample points belonging to the real label. Only if equals to , the total loss would be zero.
3.4. The Discriminator Stuck with a Classifier in CSSVGAN
As shown in
Figure 6, the Discriminator needs to specifically identify the generated data as false and the real HSI data as true. This process can be regarded as a two-category task using one-sided label smoothing: defining the real HSI data as 0.9 and the false as zero. The loss function of it marked with
is the same as the Formula (10) enumerated above. Moreover, the classifier is stuck as an interface to the output of Discriminator and the classification results are calculated directly through the SoftMax layer, where
C represents the total number of labels in training data. As mentioned above, the Encoder ensures diversity and the Generator guarantees authenticity. All these contributions place higher demands on Discriminator to achieve better classification performance. Thus, the CSSVGAN framework yields a better classification result.
The implementation details of the Discriminator in CSSVGAN are described in
Table 5 with the 3D convolution of
and strides of
. Identifying
C categories belongs to a multi-classification assignment. The SoftMax method is taken as the standard for HSIC. As shown below, the CSSVGAN method should allocate the sample
x of each class
c to the most likely one of the
C classes to get the predicted classification results. The specific formula is as follows:
Then the category of
X can be expressed as the formula below:
where
S,
C,
X,
signify the SoftMax function, the total number of categories, the input of SoftMax, and the probability that the prediction object belongs to class
C, respectively.
similar with
is a sample of one certain category. Therefore, the following formula can be used for the loss function of objective constraint.
where
n means the total number of samples,
C represents the total number of categories, and
y denotes the single label (either true or false) with the same description as above.
3.5. The Total Loss of CSSVGAN
As illustrated in
Figure 3, up till now, the final goal of the total loss of the CSSVGAN model can be divided into four parts: two KL divergence constraint losses and a mean-square error loss from the Encoder, two binary losses from the Generator, one binary loss from the Discriminator and one multi-classification loss from the multi classifier. The ensemble formula can be expressed as:
where
and
represent the loss between
or
and the standard normal distribution respectively in
Section 3.2.
and
signify the mean square error of
and
in
Section 3.3 separately.
calculates the mean square error between
and
. The purpose of
and
is to assume that the virtual data
and
(in
Section 3.3) are true with a value of one.
denotes that the Discriminator identifies
and
as false data with a value of zero. Finally, the
is the loss of multi classes of the classifier.