Figure 1.
Various attention modules, including (a) channel-wise attention, (b) spectral-wise attention and (c) spatial-wise attention. C, W, H and B represent the number of channels, width, height and number of bands respectively. For easier understanding, weighted channels, weighted bands, and weighted pixels are represented by different colors. GAP denotes the global average pooling and FC denotes the fully connected layer. Conv represents the convolutional layer.
Figure 1.
Various attention modules, including (a) channel-wise attention, (b) spectral-wise attention and (c) spatial-wise attention. C, W, H and B represent the number of channels, width, height and number of bands respectively. For easier understanding, weighted channels, weighted bands, and weighted pixels are represented by different colors. GAP denotes the global average pooling and FC denotes the fully connected layer. Conv represents the convolutional layer.
Figure 2.
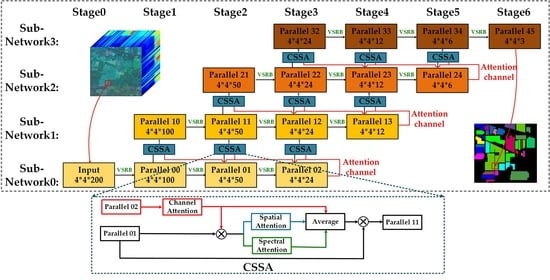
Channel–spectral–spatial-attention parallel network. Each row and column represents the same subnetwork and the same convolution stage, respectively; VSRB represents the variable spectral residual block; CSSA represents the channel–spatial–spectral-attention module.
Figure 2.
Channel–spectral–spatial-attention parallel network. Each row and column represents the same subnetwork and the same convolution stage, respectively; VSRB represents the variable spectral residual block; CSSA represents the channel–spatial–spectral-attention module.
Figure 3.
Various CNN frameworks: (a) Serial network with cross-layer connectivity, (b) network with cross-layer connectivity and multi-scale context fusion, (c) common two-branch fusion network in HSI classification, and (d) example of the proposed parallel network with two subnetworks (SN1, SN2).
Figure 3.
Various CNN frameworks: (a) Serial network with cross-layer connectivity, (b) network with cross-layer connectivity and multi-scale context fusion, (c) common two-branch fusion network in HSI classification, and (d) example of the proposed parallel network with two subnetworks (SN1, SN2).
Figure 4.
The variable spectral residual block used for replacing the convolution operation in each subnetwork. BN represents the batch normalization, and Conv denotes the convolutional layer.
Figure 4.
The variable spectral residual block used for replacing the convolution operation in each subnetwork. BN represents the batch normalization, and Conv denotes the convolutional layer.
Figure 5.
The triple-attention module, involving in channel-, spectral- and spatial- attention (CSSA). GAP, FC, K, S represents global average pooling, the fully connected layer, the kernel size and the stride, respectively. The triple-attention module(CSSA) has two inputs: (1) the feature maps at the highest level of the preceding subnetwork, and (2) the corresponding low-level feature maps of the same stage. The total weight is generated by aggregating the three types of attention modules.
Figure 5.
The triple-attention module, involving in channel-, spectral- and spatial- attention (CSSA). GAP, FC, K, S represents global average pooling, the fully connected layer, the kernel size and the stride, respectively. The triple-attention module(CSSA) has two inputs: (1) the feature maps at the highest level of the preceding subnetwork, and (2) the corresponding low-level feature maps of the same stage. The total weight is generated by aggregating the three types of attention modules.
Figure 6.
Training-test set splits: (a) random partitioning blocks, (b) training blocks, (c) leaked image, (d) remaining image, (e) validation blocks, (f) test image.
Figure 6.
Training-test set splits: (a) random partitioning blocks, (b) training blocks, (c) leaked image, (d) remaining image, (e) validation blocks, (f) test image.
Figure 7.
Training pixels in (a) Salinas Valley dataset, (b) Pavia University dataset, (c) Indian Pines dataset. Subfigure 0 in each dataset is the ground truth for the corresponding dataset. The other subfigures represent the training pixels for different folds.
Figure 7.
Training pixels in (a) Salinas Valley dataset, (b) Pavia University dataset, (c) Indian Pines dataset. Subfigure 0 in each dataset is the ground truth for the corresponding dataset. The other subfigures represent the training pixels for different folds.
Figure 8.
SerialNet and ParallelNet. VSRB represents the variable spectral residual block.
Figure 8.
SerialNet and ParallelNet. VSRB represents the variable spectral residual block.
Figure 9.
Classification maps by different models for Salinas Valley: (a) false color image, (b) ground truth, (c) SS3FCN, (d) SerialNet, (e) ParallelNet, and (f) TAP-Net.
Figure 9.
Classification maps by different models for Salinas Valley: (a) false color image, (b) ground truth, (c) SS3FCN, (d) SerialNet, (e) ParallelNet, and (f) TAP-Net.
Figure 10.
Classification maps of different models for Pavia University: (a) false color image, (b) ground truth, (c) SS3FCN, (d) SerialNet, (e) ParallelNet, and (f) TAP-Net.
Figure 10.
Classification maps of different models for Pavia University: (a) false color image, (b) ground truth, (c) SS3FCN, (d) SerialNet, (e) ParallelNet, and (f) TAP-Net.
Figure 11.
Classification maps of different models for Indian Pines: (a) false color image, (b) ground truth, (c) SS3FCN, (d) SerialNet, (e) ParallelNet, and (f) TAP-Net.
Figure 11.
Classification maps of different models for Indian Pines: (a) false color image, (b) ground truth, (c) SS3FCN, (d) SerialNet, (e) ParallelNet, and (f) TAP-Net.
Figure 12.
Classification results on none-leaked and leaked datasets for (a) Salinas Valley, (b) Pavia University, and (c) Indian Pines.
Figure 12.
Classification results on none-leaked and leaked datasets for (a) Salinas Valley, (b) Pavia University, and (c) Indian Pines.
Figure 13.
Influence of the number of labeled pixels in each block: (a) Salinas Valley, (b) Pavia University, and (c) Indian Pines.
Figure 13.
Influence of the number of labeled pixels in each block: (a) Salinas Valley, (b) Pavia University, and (c) Indian Pines.
Table 1.
Average number of training/validation/leaked/test pixels in Salinas Valley.
Table 1.
Average number of training/validation/leaked/test pixels in Salinas Valley.
| Class | Total | Train | Val | Leaked | Test | Train-Ratio (%) |
|---|
| C1 | 2009 | 77.6 | 128.4 | 605.6 | 1197.4 | 3.86 |
| C2 | 3726 | 137.6 | 90.4 | 1118.8 | 2379.2 | 3.69 |
| C3 | 1976 | 77.6 | 119 | 588.8 | 1190.6 | 3.93 |
| C4 | 1394 | 52.8 | 49 | 333.8 | 958.4 | 3.79 |
| C5 | 2678 | 102 | 118.6 | 737 | 1720.4 | 3.81 |
| C6 | 3959 | 156.4 | 147.8 | 1181.4 | 2473.4 | 3.95 |
| C7 | 3579 | 142.8 | 122.8 | 1089.6 | 2223.8 | 3.99 |
| C8 | 11,271 | 407.8 | 346 | 3509.6 | 7007.6 | 3.62 |
| C9 | 6203 | 225.8 | 275 | 1858.2 | 3844 | 3.64 |
| C10 | 3278 | 116.6 | 129 | 946 | 2086.4 | 3.56 |
| C11 | 1068 | 43.4 | 43.6 | 274.6 | 706.4 | 4.06 |
| C12 | 1927 | 75.2 | 116 | 575.6 | 1160.2 | 3.90 |
| C13 | 916 | 36.2 | 40.8 | 208.2 | 630.8 | 3.95 |
| C14 | 1070 | 44.2 | 47 | 290 | 688.8 | 4.13 |
| C15 | 7268 | 252.4 | 247.2 | 2183.2 | 4585.2 | 3.47 |
| C16 | 1807 | 68.8 | 41.6 | 571 | 1125.6 | 3.81 |
| Total | 54,129 | 2017.2 | 2062.2 | 16,071.4 | 33,978.2 | 3.73 |
Table 2.
Average number of training/validation/leaked/test pixels in Pavia University.
Table 2.
Average number of training/validation/leaked/test pixels in Pavia University.
| Class | Total | Train | Val | Leaked | Test | Train-Ratio (%) |
|---|
| C1 | 6631 | 457 | 381.6 | 1371.2 | 4421.2 | 6.89 |
| C2 | 18,649 | 1116.8 | 1332.4 | 6648.6 | 9551.2 | 5.99 |
| C3 | 2099 | 139.8 | 133.4 | 421.8 | 1404 | 6.66 |
| C4 | 3064 | 195 | 99.2 | 254.6 | 2515.2 | 6.36 |
| C5 | 1345 | 91.4 | 97.6 | 270.6 | 885.4 | 6.89 |
| C6 | 5029 | 299 | 366.4 | 1922 | 2441.6 | 5.95 |
| C7 | 1330 | 86.2 | 121.2 | 386.8 | 735.8 | 6.48 |
| C8 | 3682 | 272.6 | 164.2 | 455.2 | 2790 | 7.4 |
| C9 | 947 | 63.6 | 62.6 | 91.2 | 729.6 | 6.72 |
| Total | 42,776 | 2721.4 | 2758.6 | 11,822 | 25,474 | 6.36 |
Table 3.
Average number of training/validation/leaked/test pixels in Indian Pines.
Table 3.
Average number of training/validation/leaked/test pixels in Indian Pines.
| Class | Total | Train | Val | Leaked | Test | Train-Ratio (%) |
|---|
| C1 | 46 | 7.6 | 7.8 | 16.6 | 14 | 16.52 |
| C2 | 1428 | 159.6 | 164 | 441.4 | 663 | 11.18 |
| C3 | 830 | 97.6 | 91 | 262.2 | 379.2 | 11.76 |
| C4 | 237 | 29.4 | 24.8 | 72.2 | 110.6 | 12.41 |
| C5 | 483 | 56.2 | 52.2 | 161.4 | 213.2 | 11.64 |
| C6 | 730 | 87 | 74 | 220.8 | 348.2 | 11.93 |
| C7 | 28 | 7.2 | 8.6 | 8.4 | 3.8 | 25.71 |
| C8 | 478 | 56.8 | 55.2 | 163.8 | 202.2 | 11.88 |
| C9 | 20 | 4.6 | 3.4 | 6.6 | 5.4 | 23 |
| C10 | 972 | 122.8 | 123.8 | 330.6 | 394.8 | 12.63 |
| C11 | 2455 | 259.2 | 245.6 | 810.8 | 1139.4 | 10.56 |
| C12 | 593 | 76.4 | 63.6 | 192.4 | 260.6 | 12.88 |
| C13 | 205 | 26.6 | 31.4 | 62 | 85 | 12.98 |
| C14 | 1265 | 146 | 134.6 | 427.2 | 557.2 | 11.54 |
| C15 | 386 | 39.6 | 51 | 108.6 | 186.8 | 10.26 |
| C16 | 93 | 11.2 | 13.4 | 33.4 | 35 | 12.04 |
| Total | 10,249 | 1187.8 | 1144.4 | 3318.4 | 4598.4 | 11.59 |
Table 4.
Network framework details.
Table 4.
Network framework details.
| Stage | Stage0 | Stage1 | Stage2 | Stage3 |
|---|
| Sub-Network3: | Kernel Size | / | / | / | 3×3×3 |
| | Feature Size | / | / | / | 10 × 10 ×24 × 128 |
| Sub-Network2: | Kernel Size | / | / | 3×3×3 | 1×1×3 |
| | Feature Size | / | / | 10×10×50×64 | 10×10×24×128 |
| Sub-Network1: | Kernel Size | / | 3×3×3 | 1×1×3 | 1×1×3 |
| | Feature Size | / | 10×10×100×64 | 10×10×50×64 | 10×10×24×128 |
| Sub-Network0: | Kernel Size | 3×3×3 (Input) | 1×1×3 | 1×1×3 | 1×1×3 |
| | Feature Size | 10×10×204×1 | 10×10×100×64 | 10×10×50×64 | 10×10×24×128 |
| Stage | Stage4 | Stage5 | Stage6 | |
| Sub-Network3: | Kernel Size | 1×1×3 | 1×1×3 | 1×1×3 (Output) | |
| | Feature Size | 10×10×12×128 | 10×10×6×256 | 10×10×3×256 | |
| Sub-Network2: | Kernel Size | 1×1×3 | 1×1×3 | / | |
| | Feature Size | 10×10×12×128 | 10×10×6×256 | / | |
| Sub-Network1: | Kernel Size | 1×1×3 | / | / | |
| | Feature Size | 10×10×12×128 | / | / | |
| Sub-Network0: | Kernel Size | / | / | / | |
| | Feature Size | / | / | / | |
Table 5.
Classification results for the Salinas Valley dataset, including per-class, overall (OA), and average (AA) accuracy (in %), and the Kappa scores.
Table 5.
Classification results for the Salinas Valley dataset, including per-class, overall (OA), and average (AA) accuracy (in %), and the Kappa scores.
| Class | Method |
|---|
| VHIS | DA-VHIS | AutoCNN | SS3FCN | SerialNet | ParallelNet | TAP-Net |
|---|
| C1 | 85.91 | 96.36 | 96.75 | 92.36 | 97.88 | 99.36 | |
| C2 | 73.88 | 94.71 | 99.26 | 92.58 | 99.68 | 97.58 | |
| C3 | 33.72 | 49.95 | 79.46 | 66.35 | 90.79 | 97.78 | |
| C4 | 65.92 | 79.62 | 99.09 | 98.13 | 98.91 | 97.78 | |
| C5 | 46.42 | 64.3 | 97.21 | 95.63 | 95.34 | 94.46 | |
| C6 | 79.63 | 79.89 | 99.68 | 99.30 | 99.31 | 99.27 | |
| C7 | 73.59 | 79.62 | 99.35 | 99.43 | 99.26 | 99.43 | |
| C8 | 72.16 | 74.54 | 75.82 | 69.27 | 81.83 | 84.07 | |
| C9 | 71.87 | 96.1 | 99.05 | 99.67 | 96.84 | 98.07 | |
| C10 | 73.11 | 87.28 | 87.54 | 84.07 | 86.78 | 89.60 | |
| C11 | 72.51 | 73.08 | 89.15 | 85.31 | 73.09 | 95.38 | |
| C12 | 71.06 | 98.25 | 96.99 | 97.98 | 98.30 | 98.28 | |
| C13 | 75.80 | 97.67 | 98.36 | 98.45 | 97.22 | 97.58 | |
| C14 | 72.04 | 88.07 | 90.61 | 87.32 | 93.12 | 96.01 | |
| C15 | 45.03 | 62.92 | 63.47 | 52.31 | 50.59 | 58.84 | |
| C16 | 22.54 | 45.39 | 89.26 | 59.97 | 91.14 | 86.56 | |
| OA | 64.20 | 77.52 | 87.15 | 81.32 | 86.66 | 88.94 | 90.31 ± 1.27 |
| AA | 64.70 | 79.24 | 91.32 | 86.13 | 90.63 | 92.73 | 93.18 ± 1.27 |
| Kappa | / | 0.749 | 0.857 | / | 0.818 | 0.846 | 0.881 ± 0.03 |
Table 6.
Classification results for the Pavia University dataset, including per-class, overall (OA), and average (AA) accuracy (in %), and the Kappa scores.
Table 6.
Classification results for the Pavia University dataset, including per-class, overall (OA), and average (AA) accuracy (in %), and the Kappa scores.
| Class | Method |
|---|
| VHIS | DA-VHIS | AutoCNN | SS3FCN | SerialNet | ParallelNet | TAP-Net |
|---|
| C1 | 93.40 | 93.42 | 83.40 | 97.48 | 92.89 | 94.29 | |
| C2 | 86.20 | 86.52 | 93.32 | 90.86 | 95.40 | 93.97 | |
| C3 | 47.58 | 46.88 | 61.52 | 58.75 | 61.35 | 69.81 | |
| C4 | 86.89 | 92.21 | 78.86 | 84.81 | 81.68 | 85.31 | |
| C5 | 59.81 | 59.74 | 98.25 | 94.82 | 99.41 | 99.50 | |
| C6 | 27.14 | 27.68 | 73.34 | 23.59 | 56.58 | 71.24 | |
| C7 | 0 | 0 | 64.56 | 61.61 | 32.51 | 50.97 | |
| C8 | 78.46 | 78.32 | 76.86 | 88.84 | 69.66 | 73.52 | |
| C9 | 79.27 | 79.60 | 97.69 | 88.68 | 99.40 | 92.44 | |
| OA | 73.26 | 73.84 | 84.63 | 79.89 | 83.46 | 86.30 | 91.64 ± 1.08 |
| AA | 62.08 | 62.71 | 80.87 | 76.60 | 76.54 | 82.01 | 87.45 ± 3.09 |
| Kappa | / | 0.631 | 0.800 | / | 0.851 | 0.866 | 0.892 ± 0.02 |
Table 7.
Classification results for the Indian Pines dataset, including per-class, overall (OA), and average (AA) accuracy (in %), and the Kappa scores.
Table 7.
Classification results for the Indian Pines dataset, including per-class, overall (OA), and average (AA) accuracy (in %), and the Kappa scores.
| Class | Method |
|---|
| VHIS | DA-VHIS | AutoCNN | SS3FCN | SerialNet | ParallelNet | TAP-Net |
|---|
| C1 | 17.68 | 15.89 | 19.58 | 40.4 | 38.23 | 13.30 | |
| C2 | 56.89 | 70.41 | 60.16 | 77.89 | 57.8 | 70.86 | |
| C3 | 51.55 | 61.44 | 44.12 | 60.74 | 45.02 | 60.08 | |
| C4 | 36.27 | 42.28 | 25.35 | 11.8 | 44.1 | 53.67 | |
| C5 | 69.02 | 73.02 | 77.80 | 67.5 | 69.39 | 64.97 | |
| C6 | 92.35 | 92.13 | 90.99 | 91.95 | 92.16 | 89.06 | |
| C7 | 0 | 0 | 35.63 | 20.14 | 21.00 | 93.33 | |
| C8 | 86.95 | 86.44 | 95.87 | 81.71 | 96.77 | 98.49 | |
| C9 | 19.55 | 21.28 | 5.31 | 31.67 | 16.67 | 86.17 | |
| C10 | 60.05 | 67.47 | 55.93 | 78.15 | 64.26 | 81.34 | |
| C11 | 74.05 | 65.24 | 68.73 | 69.32 | 73.12 | 75.90 | |
| C12 | 43.71 | 49.56 | 36.96 | 40.81 | 46.97 | 71.70 | |
| C13 | 94.15 | 96.01 | 87.33 | 93.43 | 94.28 | 98.15 | |
| C14 | 91.18 | 92.68 | 84.90 | 91.77 | 91.72 | 90.04 | |
| C15 | 43.39 | 52.79 | 39.02 | 37.93 | 45.17 | 38.52 | |
| C16 | 45.04 | 44.78 | 48.02 | 75.19 | 80.79 | 80.54 | |
| OA | 67.11 | 69.49 | 65.35 | 71.47 | 69.47 | 76.59 | 81.35 ± 1.53 |
| AA | 55.11 | 58.15 | 54.73 | 60.65 | 61.09 | 73.51 | 78.85 ± 3.18 |
| Kappa | / | 0.653 | 0.600 | / | 0.695 | 0.730 | 0.787 ± 0.02 |
Table 8.
Effect of the block-patch size on OA & AA (%): (a) Salinas Valley, (b) Pavia University, and (c) Indian Pines.
Table 8.
Effect of the block-patch size on OA & AA (%): (a) Salinas Valley, (b) Pavia University, and (c) Indian Pines.
| (a) Salinas Valley |
|---|
| Block_Patch | 6_4 | 8_6 | 10_8 | 12_10 | 14_12 |
| Accuracy (%) | OA | 88.77 | 89.1 | 90.31 | 89.47 | 86.39 |
| AA | 91.9 | 93.06 | 93.18 | 93.77 | 89.4 |
| (b) Pavia University |
| Block_Patch | 6_4 | 8_6 | 10_8 | 12_10 | 14_12 |
| Accuracy (%) | OA | 88.42 | 89.41 | 91.64 | 86.38 | 83.32 |
| AA | 84.1 | 84.75 | 87.45 | 80.72 | 77.05 |
| (c) Indian Pines |
| Block_Patch | 2_1 | 4_2 | 6_4 | 8_6 | 10_8 |
| Accuracy (%) | OA | 74.21 | 76.33 | 81.35 | 79.18 | / |
| AA | 68.52 | 70.64 | 78.85 | 65.38 | / |
Table 9.
Comparison between different attention mechanisms over Pavia University dataset in terms of Per-class, overall (OA), average (AA) accuracy (in %) and the Kappa score.
Table 9.
Comparison between different attention mechanisms over Pavia University dataset in terms of Per-class, overall (OA), average (AA) accuracy (in %) and the Kappa score.
| Class | Method |
|---|
| ParallelNet | ParallelNet-SS | ParallelNet-CS | TAP-Net |
|---|
| C1 | | | | 95.67 ± 1.43 |
| C2 | | | 98.25 ± 1.69 | |
| C3 | | 85.48 ± 15.35 | | |
| C4 | | | | 94.23 ± 1.38 |
| C5 | | | 99.70 ± 0.26 | |
| C6 | | | | 84.17 ± 10.26 |
| C7 | | | 67.68 ± 19.86 | |
| C8 | | | | 83.60 ± 7.42 |
| C9 | | | | 99.33 ± 0.44 |
| OA | | | | 91.64 ± 1.08 |
| AA | | | | 87.45 ± 3.09 |
| Kappa | 0.846 ±0.01 | 0.857 ± 0.05 | 0.879 ± 0.02 | 0.892 ± 0.02 |
Table 10.
Results of two-tailed Wilcoxon’s tests over per-class accuracy for proposed methods.
Table 10.
Results of two-tailed Wilcoxon’s tests over per-class accuracy for proposed methods.
| | ParallelNet | ParallelNet-SS | ParallelNet-CS | TAP-Net |
|---|
| SerialNet | 0.007 | | | |
| ParallelNet | | 0.018 | 0.025 | 0.005 |
| ParallelNet-SS | | | 0.912 | 0.028 |
| ParallelNet-CS | | | | 0.016 |
Table 11.
Physicochemical characteristics of some grains of sorghum sampled in the main markets of Maroua town and used for the production of the indigenous beers.
Table 11.
Physicochemical characteristics of some grains of sorghum sampled in the main markets of Maroua town and used for the production of the indigenous beers.
| | | Salinas Valley | Pavia University | Indian Pines |
|---|
| | | OA | AA | Kappa | OA | AA | Kappa | OA | AA | Kappa |
| Data Splits | VHIS-VHIS | 64.2 | 64.7 | / | 73.26 | 62.08 | / | 67.11 | 55.11 | / |
| VHIS-TAP | 69.91 | 71.93 | 0.666 | 73.60 | 63.14 | 0.645 | 70.91 | 59.24 | 0.668 |
| TAP-VHIS | 85.57 | 89.04 | 0.850 | 84.60 | 81.72 | 0.805 | 57.61 | 53.15 | 0.507 |
| TAP-TAP | 90.31 | 93.18 | 0.881 | 91.64 | 87.45 | 0.892 | 81.35 | 78.85 | 0.787 |
| Data augmentation | DA-VHIS | 77.52 | 79.24 | 0.749 | 73.84 | 62.71 | 0.631 | 69.49 | 58.15 | 0.653 |
| TAP(DA) | 85.19 | 89.32 | 0.834 | 84.08 | 76.71 | 0.8 | 74.36 | 68.38 | 0.707 |
| TAP(NoDA) | 90.31 | 93.18 | 0.881 | 91.64 | 87.45 | 0.892 | 81.35 | 78.85 | 0.787 |