Assessing the Impacts of Species Composition on the Accuracy of Mapping Chlorophyll Content in Heterogeneous Ecosystems
Abstract
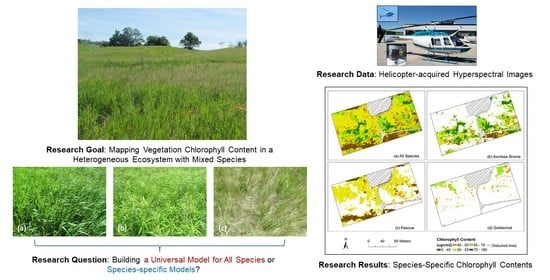
:1. Introduction
2. Material and Methods
2.1. Study Area
2.2. Field Data Collection
2.3. Collection and Processing of Hyperspectral Imagery
2.4. Establishment of Different Models for Estimating Chlorophyll
2.5. Object-Based Species Classification and Chlorophyll Mapping
3. Results and Discussion
3.1. Data Distribution Patterns of Different Species
3.2. Comparison of Different Modelling Approaches for Estimating Chlorophyll
3.3. Object-Based Species Classification
3.4. Mapping Species-Specific Chlorophyll Contents
4. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, T.; Shang, J.; Chen, J.M.; Liu, J.; Qian, B.; Ma, B.; Morrison, M.J.; Zhang, C.; Liu, Y.; Shi, Y.; et al. Assessment of Portable Chlorophyll Meters for Measuring Crop Leaf Chlorophyll Concentration. Remote Sens. 2019, 11, 2706. [Google Scholar] [CrossRef] [Green Version]
- Shang, J.; Liu, J.; Ma, B.; Zhao, T.; Jiao, X.; Geng, X.; Huffman, T.; Kovacs, J.M.; Walters, D. Mapping spatial variability of crop growth conditions using RapidEye data in Northern Ontario, Canada. Remote Sens. Environ. 2015, 168, 113–125. [Google Scholar] [CrossRef]
- Moharana, S.; Dutta, S. Spatial variability of chlorophyll and nitrogen content of rice from hyperspectral imagery. ISPRS J. Photogramm. 2016, 122, 17–29. [Google Scholar] [CrossRef]
- Wu, C.; Han, X.; Niu, Z.; Dong, J. An evaluation of EO-1 hyperspectral Hyperion data for chlorophyll content and leaf area index estimation. Int. J. Remote Sens. 2010, 31, 1079–1086. [Google Scholar] [CrossRef]
- Haboudane, D.; Tremblay, N.; Miller, J.R.; Vigneault, P. Remote estimation of crop chlorophyll content using spectral indices derived from hyperspectral data. IEEE T. Geosci. Remote 2008, 46, 423–437. [Google Scholar] [CrossRef]
- Lemaire, G.; Francois, C.; Soudani, K.; Berveiller, D.; Pontailler, J.; Breda, N.; Genet, H.; Davi, H.; Dufrene, E. Calibration and validation of hyperspectral indices for the estimation of broadleaved forest leaf chlorophyll content, leaf mass per area, leaf area index and leaf canopy biomass. Remote Sens. Environ. 2008, 112, 3846–3864. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Zhang, Y.; Simic, A.; Noland, T.L.; Nesbitt, N.; Arabian, J. Evaluating leaf chlorophyll content prediction from multispectral remote sensing data within a physically-based modelling framework. ISPRS J. Photogramm. 2015, 102, 85–95. [Google Scholar] [CrossRef]
- Croft, H.; Arabian, J.; Chen, J.M.; Shang, J.; Liu, J. Mapping within-field leaf chlorophyll content in agricultural crops for nitrogen management using Landsat-8 imagery. Precis. Agric. 2019, 21, 856–880. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Zhang, J.; Chen, D.; Huang, Y.; Kong, W.; Yuan, L.; Ye, H.; Huang, W. Assessment of Leaf Chlorophyll Content Models for Winter Wheat Using Landsat-8 Multispectral Remote Sensing Data. Remote Sens. 2020, 12, 2574. [Google Scholar] [CrossRef]
- Yu, F.; Xu, T.; Du, W.; Ma, H.; Zhang, G.; Chen, C. Radiative transfer models (RTMs) for field phenotyping inversion of rice based on UAV hyperspectral remote sensing. Int. J. Agric. Biol. Eng. 2017, 10, 150–157. [Google Scholar]
- Croft, H.; Chen, J.M.; Zhang, Y.; Simic, A. Modelling leaf chlorophyll content in broadleaf and needle leaf canopies from ground, CASI, Landsat TM 5 and MERIS reflectance data. Remote Sens. Environ. 2013, 133, 128–140. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Zhang, Y. The applicability of empirical vegetation indices for determining leaf chlorophyll content over different leaf and canopy structures. Ecol. Complex. 2014, 17, 119–130. [Google Scholar] [CrossRef]
- Simic, A.; Chen, J.M.; Noland, T.L. Retrieval of forest chlorophyll content using canopy structure parameters derived from multi-angle data: The measurement concept of combining nadir hyperspectral and off-nadir multispectral data. Int. J. Remote Sens. 2011, 32, 5621–5644. [Google Scholar] [CrossRef]
- Wong, K.K.; He, Y. Estimating grassland chlorophyll content using remote sensing data at leaf, canopy, and landscape scales. Can. J. Remote Sens. 2013, 39, 155–166. [Google Scholar] [CrossRef]
- Lu, B.; He, Y. Evaluating Empirical Regression, Machine Learning, and Radiative Transfer Modelling for Estimating Vegetation Chlorophyll Content Using Bi-Seasonal Hyperspectral Images. Remote Sens. 2019, 11, 1979. [Google Scholar] [CrossRef] [Green Version]
- Lu, B.; He, Y.; Liu, H.H.T. Mapping vegetation biophysical and biochemical properties using unmanned aerial vehicles-acquired imagery. Int. J. Remote Sens. 2018, 39, 5265–5287. [Google Scholar] [CrossRef]
- Guo, C.; Guo, X. Estimating leaf chlorophyll and nitrogen content of wetland emergent plants using hyperspectral data in the visible domain. Spectrosc. Lett. 2016, 49, 180–187. [Google Scholar] [CrossRef]
- Zhuo, W.; Shi, R.; Wu, N.; Zhang, C.; Tian, B. Spectral response and the retrieval of canopy chlorophyll content under interspecific competition in wetlands—case study of wetlands in the Yangtze River Estuary. Earth Sci. Inform. 2021, 14, 1467–1486. [Google Scholar] [CrossRef]
- Kanning, M.; Kuehling, I.; Trautz, D.; Jarmer, T. High-Resolution UAV-Based Hyperspectral Imagery for LAI and Chlorophyll Estimations from Wheat for Yield Prediction. Remote Sens. 2018, 10, 2000. [Google Scholar] [CrossRef] [Green Version]
- Darvishzadeh, R.; Atzberger, C.; Skidmore, A.; Schlerf, M. Retrieval of vegetation biochemicals using a radiative transfer model and hyperspectral data. In Proceedings of the ISPRS Technical Commission VII Symposium—100 Years ISPRS Advancing Remote Sensing Science, Vienna, Austria, 5–7 July 2010; Volume 38, pp. 171–175. [Google Scholar]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; Francois, C.; Ustin, S.L. PROSPECT plus SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Lu, B.; He, Y. Leaf Area Index Estimation in a Heterogeneous Grassland Using Optical, SAR, and DEM Data. Can. J. Remote Sens. 2019, 45, 618–633. [Google Scholar] [CrossRef]
- Historical Climate Data. Available online: http://climate.weather.gc.ca/ (accessed on 15 September 2021).
- Minocha, R.; Martinez, G.; Lyons, B.; Long, S. Development of a standardized methodology for quantifying total chlorophyll and carotenoids from foliage of hardwood and conifer tree species. Can. J. Forest Res. 2009, 39, 849–861. [Google Scholar] [CrossRef]
- Wong, K.K.L. Remote Sensing pf Tall Grasslands: Estimating Vegetation Biochemical Contents at Multiple Spatial Scales and Investigating Vegetation Temporal Response to Climate Conditions. Master’s Thesis, University of Toronto, Toronto, ON, Canada, 2013. [Google Scholar]
- Lucieer, A.; Malenovský, Z.; Veness, T.; Wallace, L. HyperUAS-imaging spectroscopy from a multirotor unmanned aircraft system. J. Field Robot. 2014, 31, 571–590. [Google Scholar] [CrossRef] [Green Version]
- Lu, B.; Proctor, C.; He, Y. Investigating different versions of PROSPECT and PROSAIL for estimating spectral and biophysical properties of photosynthetic and non-photosynthetic vegetation in mixed grasslands. GIScience Remote Sens. 2021, 58, 354–371. [Google Scholar] [CrossRef]
- Dao, P.D.; He, Y.; Lu, B. Maximizing the quantitative utility of airborne hyperspectral imagery for studying plant physiology: An optimal sensor exposure setting procedure and empirical line method for atmospheric correction. Int. J. Appl. Earth Obs. 2019, 77, 140–150. [Google Scholar] [CrossRef]
- Lu, B.; He, Y.; Dao, P.D. Comparing the Performance of Multispectral and Hyperspectral Images for Estimating Vegetation Properties. IEEE J. STARS 2019, 12, 1784–1797. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, O.B.; Singh, B.R. A comparative assessment of support vector regression, artificial neural networks, and random forests for predicting and mapping soil organic carbon stocks across an Afromontane landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Main, R.; Cho, M.A.; Mathieu, R.; O’Kennedy, M.M.; Ramoelo, A.; Koch, S. An investigation into robust spectral indices for leaf chlorophyll estimation. ISPRS J. Photogramm. 2011, 66, 751–761. [Google Scholar] [CrossRef]
- Tong, A.; He, Y. Estimating and mapping chlorophyll content for a heterogeneous grassland: Comparing prediction power of a suite of vegetation indices across scales between years. ISPRS J. Photogramm. 2017, 126, 146–167. [Google Scholar] [CrossRef]
- Spectral Indices. Available online: https://www.harrisgeospatial.com/docs/AlphabeticalListSpectralIndices.html (accessed on 10 December 2018).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. Mach. Learn. 2011, 12, 2825–2830. [Google Scholar]
- Yu, X.; Hyyppä, J.; Vastaranta, M.; Holopainen, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on the random forests technique. ISPRS J. Photogramm. 2011, 66, 28–37. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using WorldView-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Abdel Rahman, E.M.; Ismail, R. Estimating standing biomass in papyrus (Cyperus papyrus L.) swamp: Exploratory of in situ hyperspectral indices and random forest regression. Int. J. Remote Sens. 2014, 35, 693–714. [Google Scholar] [CrossRef]
- Karlson, M.; Ostwald, M.; Reese, H.; Sanou, J.; Tankoano, B.; Mattsson, E. Mapping tree canopy cover and aboveground biomass in sudano-sahelian woodlands using Landsat 8 and random forest. Remote Sens. 2015, 7, 10017–10041. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Rahman, E.M.; Ahmed, F.B.; Ismail, R. Random forest regression and spectral band selection for estimating sugarcane leaf nitrogen concentration using EO-1 Hyperion hyperspectral data. Int. J. Remote. Sens. 2013, 34, 712–728. [Google Scholar] [CrossRef]
- Lu, B.; He, Y. Species classification using Unmanned Aerial Vehicle (UAV)-acquired high spatial resolution imagery in a heterogeneous grassland. ISPRS J. Photogramm. 2017, 128, 73–85. [Google Scholar] [CrossRef]
- Lebourgeois, V.; Dupuy, S.; Vintrou, E.; Ameline, M.; Butler, S.; Begue, A. A Combined Random Forest and OBIA Classification Scheme for Mapping Smallholder Agriculture at Different Nomenclature Levels Using Multisource Data (Simulated Sentinel-2 Time Series, VHRS and DEM). Remote Sens. 2017, 9, 259. [Google Scholar] [CrossRef] [Green Version]
- Cho, M.A.; Skidmore, A.; Corsi, F.; van Wieren, S.E.; Sobhan, I. Estimation of green grass/herb biomass from airborne hyperspectral imagery using spectral indices and partial least squares regression. Int. J. Appl. Earth Obs. 2007, 9, 414–424. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Tulpan, D.; Eskandari, M. Using Hybrid Artificial Intelligence and Evolutionary Optimization Algorithms for Estimating Soybean Yield and Fresh Biomass Using Hyperspectral Vegetation Indices. Remote Sens. 2021, 13, 2555. [Google Scholar] [CrossRef]
- Liakos, K.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Types | Predictor Variables |
|---|---|
| Reflectance (nm) | Re400, Re420, Re440, Re460, Re480, Re500 … Re960, Re980, Re1000 |
| Principal Components | PC1, PC2, PC3, PC4, PC5 |
| Spectral Indices [16,31,35] | Normalized Difference Vegetation Index (NDVI) Simple Ratio Index (SRI) Enhanced Vegetation Index (EVI) Atmospherically Resistant Vegetation Index (ARVI) Red Edge Normalized Difference Vegetation Index (RENDVI) Modified Red Edge Simple Ratio Index (MRESRI) Modified Red Edge Normalized Difference Vegetation Index (MRENDVI) Sum Green Index (SGI) Vogelmann Red Edge Index 1 (VREI1) Vogelmann Red Edge Index 2 (VREI2) Vogelmann Red Edge Index 3 (VREI3) Red Edge Position Index (REPI) Photochemical Reflectance Index (PRI) Structure Insensitive Pigment Index (SIPI) Red Green Ratio Index (RGRI) Plant Senescence Reflectance Index (PSRI) Carotenoid Reflectance Index 1 (CRI1) Carotenoid Reflectance Index 2 (CRI2) Anthocyanin Reflectance Index 1 (ARI1) Anthocyanin Reflectance Index 1 (ARI2) Water Band Index (WBI) |
| Texture Metrics | b450-Mean, b450-Variance, b450-Homogeneity, b450-Contrast, b450-Dissimilarity, b450-Entropy, b450-Second Moment, b450-Correlation b550-… b670-… b680-… b704-… b750-… b800-… |
| Top 10 Important Variables in the Species-Specific Models | ||||
|---|---|---|---|---|
| Also Top 10 Important in the Universal Model | NOT Top 10 Important in the Universal Model | |||
| Awnless Brome Model | VREI1 b680-Mean RENDVI MRENDVI b670-Mean | MRESRI Re680 Re660 NDVI | CRI1 | |
| Fescue Model | RENDVI VREI1 RGRI | SIPI PC3 b680-Correlation b670-Correlation | WBI ARVI CRI1 | |
| Goldenrod Model | RGRI NDVI | Re740 PC1 Re900 b750-Mean | Re940 EVI Re920 Re800 | |
| Classified | ||||||
|---|---|---|---|---|---|---|
| Awnless Brome | Fescue | Goldenrod | All | Producer’s Accuracy | ||
| Actual | Awnless Brome | 171 | 1 | 4 | 176 | 97% |
| Fescue | 0 | 60 | 1 | 61 | 98% | |
| Goldenrod | 8 | 5 | 43 | 56 | 77% | |
| All | 179 | 66 | 48 | 293 | - | |
| User’s Accuracy | 96% | 91% | 90% | - | 94% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, B.; He, Y. Assessing the Impacts of Species Composition on the Accuracy of Mapping Chlorophyll Content in Heterogeneous Ecosystems. Remote Sens. 2021, 13, 4671. https://doi.org/10.3390/rs13224671
Lu B, He Y. Assessing the Impacts of Species Composition on the Accuracy of Mapping Chlorophyll Content in Heterogeneous Ecosystems. Remote Sensing. 2021; 13(22):4671. https://doi.org/10.3390/rs13224671
Chicago/Turabian StyleLu, Bing, and Yuhong He. 2021. "Assessing the Impacts of Species Composition on the Accuracy of Mapping Chlorophyll Content in Heterogeneous Ecosystems" Remote Sensing 13, no. 22: 4671. https://doi.org/10.3390/rs13224671
APA StyleLu, B., & He, Y. (2021). Assessing the Impacts of Species Composition on the Accuracy of Mapping Chlorophyll Content in Heterogeneous Ecosystems. Remote Sensing, 13(22), 4671. https://doi.org/10.3390/rs13224671