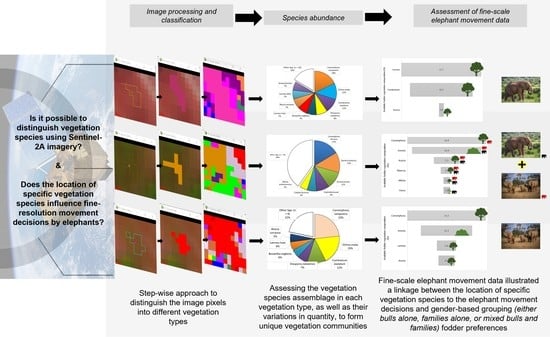
1. Introduction
Historically, elephants and humans have been integral players in the structuring of the African savanna landscape [
1]. However, this has dramatically changed in recent times, with human practices playing a more significant role as the dominant disruptive agents with unforeseen consequences to the ecosystem and biodiversity function [
2]. Increasing human populations have resulted in the rapid expansion of human infrastructure in road networks, settlements, and cultivated fields into previously wildlife natural habitats. Consequently, there has been increasing loss of natural wildlife habitat, compromised landscape connectivity, degraded natural fodder and ultimately, the elephant populations have significantly declined in relation to their historical ranges and sizes [
3,
4]. As their natural habitat diminishes, elephants and people are progressively in closer contact with each other, resulting in what is termed Human-Elephant Conflict (HEC) [
5]. HEC constitutes conflict over shared resources and space, and has detrimental consequences of crop raiding and destruction, and, in extreme instances, loss of life [
6,
7,
8,
9,
10]. Continued landscape transformations by the increasing human populations pushes elephants and humans to live in closer proximity, thereby contributing to increased conflict cases, with significant fatal incidences [
5]. In Kenya alone, it is reported that 100 problem elephants on average are shot by wildlife authorities, and about 200 cases were reported of elephant conflict related deaths between 2010 and 2017 [
11]. The intensity of crop-raiding in farmland areas within 5 km from national parks’ and protected areas’ boundaries has been on the rise since 2014 [
12] as a result of increased drought periods and reduced rainfall which lowers the quality of natural forage, hence making crops, at all growth stages, the next likely fodder option [
13,
14,
15,
16]. Studies conducted in two regions of varying vegetative climatic conditions showed significant economic losses to crop-raiding elephants; 414 farmers in the semi-arid areas around the Marsabit National Park and Reserve made loses amounting to USD 208,814 within the period of a year, between August 2004 and July 2005 [
17], averaging about USD 500 per farmer, while their counterparts farming in the highland areas bordering Meru National Park incurred more loses to the tune of USD 120,308 amongst 144 farmers, averaging at USD 835 per farmer within a similar one-year time frame, between August 2010 and July 2011 [
18]. Conflict in other parts of the world is documented in Sri Lanka, which is home to approximately 3000 elephants, and documents over 200 elephant and 70 human mortalities annually from human-elephant conflict [
19,
20]. In India, the deaths of 100 elephants and 400 humans have been recorded as a result of conflict incidences, as well as direct afflictions to over 500,000 families as a result of crop-raiding [
21]. HEC has become a considerable threat to the conservation of biodiversity, and thus it is the goal of afflicted countries to manage this conflict. There has been considerable growth in the understanding of elephant behavior, as well as the spatial-temporal patterns of HEC, and this has in turn lead to the proposal, development, implementation and reproduction of a wide array of deterrence and mitigation approaches [
13,
22,
23,
24,
25].
Kenya presents common conflict instances between elephants that are free-ranging and farmers. One such conflict zone is the community area of Sagalla, situated between the key Tsavo East and West national parks, which are together home to 14,964 elephants based on the 2021 national wildlife census [
26]. Key consequences of this human expansion in the Sagalla area have seen an increase in elephant crop-raids, breakage of farm storage houses and water tanks to access food and water resources [
25]. Deforestation for this cultivation expansion has also contributed to reduced soil moisture content, prolonged drought periods, reduced vegetation species diversity through habitat fragmentation and negative activities such as a reliance on charcoal production [
27,
28].
How plant diversity affects elephant movement patterns on a macro scale is less understood. The extent of research conducted to date on vegetation-related influences on elephant movement and presence include; a study of elephant response to vegetation spatial heterogeneity as well as patch size [
29], elephant movement in response to precipitation-driven dynamics of vegetation [
30], the change of vegetation in landscapes frequented by elephants [
31,
32,
33,
34], the seasonal preferences of elephants across wet and dry savanna landscapes where they found that elephants consistently seek outgreen vegetation patches all year round [
35]. There has been no study investigating the location of specific vegetation species as a driver of elephant movement or presence. The closest to this was an investigation of vegetation species utilized by elephants, and the specific preferences by bull elephants versus family groups [
36,
37,
38]. In addition, obtaining updated quality vegetation data for landscapes and buffer zones where elephants reside is a significant necessity for conservation planning. This is the case because an increase in the quantity and quality of data is crucial to fill gaps in scientific knowledge pertinent to enhancing conservation forecasts [
39,
40]. Knowledge gaps in species taxonomy hinder progress in understanding vegetation distribution, abundance, evolutionary patterns, biotic and abiotic interactions and traits, respectively, [
41], altogether impeding knowledge gain in ecological functionality, and ultimately introducing uncertainties in planning for conservation and management due to data insufficiencies [
40]. In this study, we propose using a vegetation community mapping method to attempt to classify the vegetation buffer zone in an area where elephants frequently stage raids into a neighboring farming community from Tsavo East National Park in southern Kenya [
25]. Understanding resource utilization of natural vegetation by elephants outside of National Parks will help managers better understand how to manage buffer zones and focus limited resources onto potential conflict hotspots.
The quantification of vegetation cover at local and global scales at a defined period in the development of a map provides valuable information for understanding the balance of natural and human-made environment. The appreciation of vegetation structure can reveal the qualities of the sites upon which it occurs as it is closely tied to its environment. This is in cases for instance where modeling vegetation dynamics is vital in communities that present frequent vegetation disturbance [
42], where the valuations and perceptions of urban wastelands are influenced by the structure of vegetation [
43], and where capacity building for monitoring and management of natural vegetation resources is prudent [
44] The information contained in resulting maps is applied as a tool of environmental planning and management, to fields such as forestry, nature conservation, landscape architecture, plant and animal ecology, agriculture, and climatology [
45].
Remote sensing earth observation techniques have evolved and been critical in large-scale map productions as it permits repeated and consistent assessment and monitoring of the environment by allowing independent control, with the provision for quality checks. As such, it is a tool with very desirable characteristics for supporting environmental policy [
46]. Earth observation data is beneficial as it is acquired in a variety of modes including optical, LIDAR and radar. The data from satellite sensors are acquired in multiple resolutions, bandwidth, and in varying conditions [
47]. However, ground information explaining the phenomenon being observed needs to be acquired by employing vegetation survey techniques to ensure that the interpretation of the earth observation data is accurate. This also goes to ensure that classification outputs are appropriate for actualization in conservation processes. There are common hindrances to achieving this link, often including limited knowledge of habitats by the mapping scientists, and the skepticism on the effectiveness of the system to accurately depict ground information [
48].
Sentinel-2 is the latest-generation high resolution open-source Earth observation satellite of the European Space Agency (ESA) for land and coastal applications. It is one of the Copernicus program launched in June 2015. It is aimed at continued independent global observation. Sentinel-2 provides imagery with increased spectral and spatial resolution. It has 13 spectral bands, from blue to SWIR (shortwave infrared), including red-edge bands. Its bands are at 10 m to 60 m spatial resolution. This has successfully been applied in land use and land cover mapping [
49], forest stress monitoring [
50], and a variety of land monitoring applications [
51], such as water detection and crop type and tree species identification [
52].
Researchers have intimated that for high vegetation classification accuracies to be achieved, more information beyond the spectral reflectance would be required including; measures of biophysical parameters, the structural characteristics of the forest, heterogeneity of the landscape among others, and utilized in object-based algorithms [
53,
54,
55]. This study will assess the viability of spectral-based classification for vegetation species and communities considering time and cost- effectiveness of using high resolution imagery and supporting in situ measurements, which compete with traditional survey methods [
53]. Levels of spatial segmentation and generalization of vegetation are driven by the usefulness of resulting information, which in turn varies based on the objective. The size of the territory is also a key determinant of the suitable geographic scale at which the vegetation is classified and mapped [
56]. In this research, we will be using Sentinel 2A imagery to assess its viability for mapping vegetation communities at species level using pixel-based algorithms and conducting in situ ground truthing assessments to identify the vegetation species. At this proposed large scale of classification and mapping, we will be adopting the species dominance classification criteria to define dominance-community types or floristic units [
56], based on one or more of the dominant species for the associated classification class [
57,
58]. We aim to map the vegetation communities and composition in this area to: (i) determine the natural vegetation species communities and their concentrations in the resulting classification classes, and (ii) investigate if these vegetation communities’ locations have any significance to the elephants moving in this landscape, and hence if it could be an important factor advising their movement decisions
2. Materials and Methods
2.1. Study Area
The study was conducted in the naturally vegetated area of Sagalla, south of the Sagalla hill (
Figure 1). Agricultural farming is the main source of income for communities living at the bottom of the hill. The Sagalla area is located south of the Tsavo East National Park, situated 3 km close to the park boundary, and separated from the fence by the newly developed standard gauge railway (SGR) and the busy Nairobi-Mombasa tarmacked highway. To the east and south of Sagalla beyond the populated villages, there are large areas of natural vegetation, and partitioned ranches which still boast huge expanses of naturally vegetated land. To the west of Sagalla is the Sagalla hill where more farming is practiced at the top of the hill and is devoid of human-elephant conflict. Land to the south-west of Sagalla is partitioned as ranches but with mostly natural vegetation cover and hence wildlife presence. These ranches are left open with no fences to allow the free-flow movement in and out of the ranches. Elephants migrate north and south between the parks and hence pass through the Sagalla community area where we are presented with cases of human-elephant conflict. It is in this Sagalla community area that we have documented micro-movements of elephants from the Tsavo East and West National Park boundary fences into the community area and back, raid crops and destroy farm properties such as water tanks and houses within the period 2015 to January 2020.
2.2. Product Description and Pre-Processing Techniques
Sentinel 2A is a polar orbiting, multispectral, and high-resolution imaging mission. Sentinel 2A instrumentation comprises of 13 spectral channels, captures a swath width of 290 km allowing for large scale main category mapping, and bands at a spatial resolution of 10 m (4 visible and NIR bands), 20 m (6 Red-edge/SWIR bands), 60 m (3 atmospheric correction bands). Sentinel 2A data is available for free download from ESA Copernicus website [
59]. Copernicus have also provided SNAP (Sentinel Application Platform) software from which to conduct spatial analysis on the Sentinel 2A imagery.
The downloaded product for Sentinel 2A satellite mission was product Level 1C (L1C) Copernicus Sentinel data 2017, retrieved from ASF DAAC on 26-01-2017, processed by ESA. This product’s description indicated that it had already been taken through several pre-processing stages namely; (i) telemetry analysis and decompression of the downloaded product at Level O, (ii) radiometric correction and geometric model refinement using the default 90 m SRTM DEM and global referencing images at Level 1A, (iii) resampling and conversion of pixel values to Top of Atmosphere (TOA) reflectance at Level 1B, and (iv) correction for gas and smile and water vapor retrieval, finally outputting Level 1C and cloud masks.
The radiometric correction performed on the L1C corrects for dark signal attributed to sun angle effects, pixel response non-uniformity, crosstalk, and identifies defective pixels that can be masked out ahead of processing. It also restores the high spatial resolution bands by noise removal and de-convolution. This pre-processing information is as provided for in the online Sentinel technical guide as captured in their website [
60].
As illustrated in
Figure 2, we introduced the downloaded L1C product into SNAP environment whereby the TOA reflectance passed through the Sen2Cor processor for terrain, cirrus and atmospheric to derive a sentinel L2A BOA product. We performed several alterations to the Sen2Cor configuration settings in L2A_GIPP.xml to facilitate cirrus removal and BRDF correction and defining the amount of cloud detection by setting the WV cirrus threshold to 0.25 < 0.34.
During the time of processing, sen2three product was not available to process 2017 sentinel 2A imagery. Sen2three toolkit replaces bad pixels (no data, saturated, defective, dark, cloud shadows, unclassified, medium and high probability clouds, thin cirrus and ice or snow) of an image with good ground pixels (vegetation, soil/rock, water, built surfaces) of recent or current earth observation imagery. There are a variety of algorithm options that can be determined in the configuration stages which include; (i) most recent- whereby bad pixels of the previous scene are replaced with good pixels of the most recent scene in the collection, and (ii) temp homogeneity- whereby previous scene pixels are replaced only if the sum of current scene pixels is higher than the best of the most current scene.
Failure to perform further image enhancements using this Sen2three toolkit to replace bad pixels, we masked out cloudy areas and shadows. The L2A output was resampled to 10 m spatial resolution to facilitate reprojection to UTM projected coordinate system. This output was then exported with the (.evf) extension to facilitate further processing in the ENVI environment. ENVI processing involved layer-stacking bands 2-8A,11 and 12, mosaicking, using color composites to visually pick out training regions, and supervised classification using maximum likelihood classification.
We developed comparisons between Landsat 8 and Sentinel 2A band resolutions to define suitable feature identification composite band combinations using bandwidth information as in
Table 1 and
Table 2. We compared the bandwidth positions of Sentinel 2A bands electromagnetic spectrum and cross-checked the best fit along the Landsat 8 spectrum. Sentinel 2 band 8A is specifically chosen for atmospheric applications instead of band 8 as is indicated in the Sentinel 2A user guides.
2.3. Supervised Classification
In ENVI environment, we applied false colour composite band combinations to define pixels or polygons to be used as training regions. The true colour rendition displays the red, blue, and green bands such that the output image is as close as possible to reality. Bands in the visible and NIR spectrum are used to detect photosynthetic vegetation while the SWIR contribute to separate individual contributions of non-photosynthetic and bare-soil structures. To distinguish vegetation features, a colour composite including the Near-IR band 8 is used. Green vegetation reflects Infrared light energy that is depicted on the image as the very red feature. Shades of red illustrate the different vegetation signatures on the landscape dependent on the leaf and canopy structure composition. Utilizing the Shortwave IR band 2000 nm to 2300 nm greatly and accurately improves the sub-pixel fractional covers of photosynthetic, non-photosynthetic and baresoil constituents’ estimation. Sentinel 2A provides a spectral coverage of 2100 nm to approximately 2380 nm in the Shortwave IR to carry out this level of feature distinction and extraction.
We generated Regions of Interest (ROI) to serve as the training areas upon which the cover types would be defined. The main training classes used as the benchmark for the classification define; grassland vegetation, forest vegetation, built-up areas, cultivated areas, water, wetland vegetation, shrubland, and bareland. To accommodate differences in canopy structures and standard dynamics, we branched out the training classes to create more ROIs. This was followed by conducting a measure of the separability between ROIs using the Compute ROI Separability tool option in ENVI between generated classes with close enough colour shades intensities while displaying the false colour composites. The ROI separability tool applies the Jeffries-Matusita distance and Transformed Divergence to output divergence metrics between defined classes and values it ranging from 0 to 2.0. A return of 2.0 means that the ROIs are completely separable and there is confidence in running them through the classifier. A return of metric values between 1.9 and 2.0 means that the ROIs have overall good separability and can be passed on to the classifier, but with potential of pixel misclassification if they are between 1.90 and 1.98. For values less than 1.9 it means the ROIs have fair separability, and the classes should either be merged to avoid misclassification [
61], or they should be split further if having distinctly separable spectral signatures for classification, allowing for an option of recombining them post-classification. We used the n-dimensions (n-D) visualizer tool to further validate the uniqueness of these ROI pixels. The n-D visualizer generates a spectral scatter plot, where n represents the number of bands. For our application, the ROIs were plotted in a 10-axis plots, equivalent of the 10 bands used in the classification process. The coordinates of the ROI points in the n-D space consist of 10 spectral reflectance band values for each ROI pixel. Our direct import of the ROIs to the n-D visualizer was not based on the purest image pixels, hence it is possible some endmembers would miss in the resulting scatter plot. Endmembers are considered spectrally unique pure pixels that occur in an image scene and can be generated using a linear unmixing model.
ROI feature extraction was achieved by digitizing points and polygons using the ROI tool dialogue. The generated Image, Scroll and Zoom display windows are used to pan across the landscape. The Zoom display is used to digitize the training pixels, using the Image window to instantly pan onto that same area on Google Earth imaging platform in true color display. This step is crucial to affirm cover type characteristics and make an informed decision of the large-scale vegetation structure and land use characteristics. Several training sites are developed for generated classes, with the option of branching out and creating new classes that would adequately describe the complexity of the local vegetation pattern at the 10 m spatial scale, whose distinctive difference from training pixels of a similar class is determined using the separability tool and n-d visualizer. This refines class definitions to decrease ambiguity ahead of adopting the Maximum Likelihood Classification system for this classification.
2.4. Ground Truthing for Species Identification and Verification
We collected referencing data and measurements to observe the ground occurrence of the vegetation in the period from May 2017 to June 2018. This process was both expensive and consumed a lot of personnel time capacity taking extra caution accessing sampling points in densely forested areas as well as being on the look out to avoid any contact with wildlife. We involved local and professional botanists with deep knowledge of the local vegetation, as well as security personnel during these field expeditions. The ground truthing was essential in the interpretation of each cover type phenomena. Through comparison of randomly selected sampling points, it aided in the analysis of species contribution to the spectral information. This analysis obtained the quantitative estimates relevant to class distributions. A selected number of ground truthing points of known species locations, depicting high concentrations for the specific species were used in the validation of the supervised classification accuracy level.
Since the satellite data spatial resolution used was at 10 m, we determined that we would study the present vegetation species at 10 m radius from the sampling point’s geographical location. We used A GPS Garmin eTrex 20× to get to the sampling point locations set at 3 m accuracy ensuring at least 8 satellites were locked in before recording data. Some sites were densely forested so, aware of the risk of a weakened signal and potential for error, we maintained collecting data on the species in a 10 m radius from the central GPS point so long as the GPS locked in a minimum of three satellites [
62].
We selected several sampling sites per classification class to determine species dominance. We then used this data to define the plant community based on quantifiable parameters. At each sampling site we collected species data based on percentage frequency and percentage cover. In some instances, we made computations of species density to supplement the spatial record of occurrences in places with a variety of tree species. Recorded field data comprised of:
- i.
Frequency- the number of quadrats within which an individual species appears
- ii.
Cover and Density- combined influence of the percentage of ground covered by a species determined by the size of canopy viewed form an aerial perspective, collected from the top (trees) down (shrubs), as well as a head count of the number of individual species occurring at one sampling point. Done for sampling plots with numerous numbers of tree and shrub species.
2.5. Species Dominance Characterization
This was achieved by determining the Importance Value Index (IV) of the vegetation specie composition within 10 m radius of the GPS sampling point. The IV is the measure of the dominance of a vegetation species in a sampling plot. IV is the measure of species diversity and richness, whereby the vegetation data is quantitatively analyzed for relative cover and relative frequency [
63,
64,
65]. The data collected and used to determine this includes (see
Table 3):
Relative frequency- this is the percentage of points occupied by a specific vegetation specie as a function of all species present
Relative cover/abundance/density- which is the number of individual vegetation species in an area as a percentage of the total individuals of all vegetation species
The equations used to calculate the IV for each cover type include [
66]:
From the raw species’ cover observations per ground truth site
,
is the cover or abundance of each vegetation species type where
is the individual species type.
is the total number of sample ground- truthing sites for each classification class
Cover was determined using:
where
the cover of the species
and
is the sample plot identity,
is the total cover area of a sample plot in this case documented as 1 to represent the complete 10 m radius from central GPS point location, while
the estimated score count in percentage based on the Braun-Blanquet abundance/cover scale [
67] shown in
Table 4 for species
in sample plot
, as well as an informed estimate ‘by eye’ of top-down canopy cover and tree height position influence.
We followed this with computing for the relative cover
where
all species occurring within each plot
and
is the relative cover in percentage of each individual species. The relative cover of an individual species across all ground truthing plots for each classification class was computed as
The relative frequency for each individual vegetation species within a classification class was determined using
where
is the relative frequency of an individual species
,
is the total number of plots in which the individual species
occurs, and
is the total number of sample plots used to ground truth for a specific classification class
.
The Relative Importance Value index (RIV) was determined further down the line from the Importance Value index (IV) which was the summative value out of 200% of the relative frequency and the relative cover as in
for all the vegetation species occurring in a specific classification class
This result would equal to 200% for classification class, and would then be divided by 2 to give the RIV resultant value out of 100% to represent the dominance of an individual vegetation species in the associated classification class hence:
Using the RIV dominance values per classification class, we could now proceed to assigning names of classification classes based on the floristic association technique, where the scientific names of the most dominant vegetation species are used.
2.6. Naming Floristic Association Vegetation Classification Classes
We followed the nomenclature rules and guidelines for assigning of ecological community scientific names [
34], to the vegetation classification classes for which we had sampled adequate ground truthing data. Association names require a maximum of five species which would be necessary for the classification classes with very diverse species types of considerably even dominance RIV values and varying total composition, for purposes of clarity. However, fewer species names are desirable in the final name. We assessed the top five RIV species for each classification category and determined how many species would be present in the name dependent on the spread of dominance value positions. We considered the species to be occurring in the same stratum hence the species labels were separated using
, a hyphen with spaces. Therefore, the order of species in the assigned association level name would reflect decreasing dominance level. Where the complete species name was not known, we used the general term spp. as the species placeholder, for instance
Indigofera spp. At the end of each association name, we added the general classification type such as ‘grassland’, ‘woodland’, or ‘shrubland’.
2.7. Introducing Elephant Tracking Data and Analysing the Relationship with Vegetation Communities
We incorporated the in person-collected elephant tracking data collected by our team at Save the Elephants over six years from 2015 to January 2020, a total of 268 tracks across this community area and ranch landscape (
Figure 3). Each elephant movement track applied in this study was collected the day following an elephant raid or visit event using the mobile handheld Garmin etrex GPS on foot, from a starting point within the community area or park fence and following the elephant footprints (measured for accuracy purposes) up to where the field team either loses the track or meets a barrier like the park fence, or in extreme cases, came close to the elephants they were tracking. The tracks were recorded as points in GPS Exchange format (gpx). Each point of a typical gpx track contained; an identity label, latitude and longitude coordinates, a local time stamp, and elevation data. The total movement data recorded and used in this study was about 447 km, with the shortest track being about 200 m and the longest about 30 km, and the mean of all tracks being 1.7 km. The map shows the extent of the elephant tracks across the study area landscape. They traverse the area comprised of natural vegetation and into the farms. Farm areas are excluded from this study as we only assessed the influence of natural vegetation species as a motivation to their movement patterns and location.
We used the raw point movement data to extract the classification output raster values to each point. We then performed summary statistics on the extracted data to extract information on which movement points fell within each classification class, as well associated attribute information of the gender grouping of the elephants, as well as the minimum and maximum number of elephants recorded to have followed the associated track.









































 = High acceptance
= High acceptance  = Moderate acceptance
= Moderate acceptance  = Low acceptance
= Low acceptance  = No acceptance.
= No acceptance.