In Situ Measuring Stem Diameters of Maize Crops with a High-Throughput Phenotyping Robot
Abstract
:1. Introduction
- A strategy of stem point cloud extraction is proposed to cope with the stems in the shade of dense leaves. This strategy solves the problem of extracting stem point clouds under canopy with narrow row spacing and cross-leaf occlusion;
- A real-time measurement pipeline is proposed to estimate the stem diameters. In this pipeline, we present two novel stem diameter estimation approaches based on stem point cloud geometry. Our approaches can effectively reduce the influences of depth noise or error on the estimation results;
- A post-processing approach is presented to fill the missing parts of the stem point clouds caused by the occlusion of dense adjacent leaves. This approach ensures the integrity of the stem point clouds obtained by RGB-D cameras in complex field scenarios and improves the accuracy of stem diameter estimation.
2. Related Works
2.1. HTP Platforms
2.2. HTP Robots
2.3. Phenotyping Sensors
2.4. Maize Phenotyping
3. Materials and Methods
3.1. HTP Platform
3.2. Field Data Collection
3.3. Data Processing
3.3.1. Extraction of Stem Point Cloud
3.3.2. Estimation of Stem Diameters
3.3.3. Filling Strategy for Missing Stem Parts in the Point Cloud
- (i).
- Traversing the point cloud from the minimum value to the maximum value for the Y coordinate at a grid threshold interval of 0.003 m on the Y-axis. The range of the traversal is given by:
- (ii).
- For the region between row i and row i + 1 on the Y-axis during the traversal process, if the region has 3D points, that is, Equation (7) is satisfied, the region does not need to be filled;
- (iii).
- If the requirements of the previous step are not met, point cloud filling is performed on the correspondence region;
- (iv).
- The number of points that need to be filled in this region can be expressed as:
- (v).
- The X and Y coordinates of the added points are given by:
- (vi).
- The Z values of the points are set to 0 s. The reason is that the approach of calculating the stem diameters by SD-PPC does not need the Z values. Meanwhile, for the SD-PCCH approach, the convex hull already encloses the missing point cloud area, so there is also no need to calculate the Z values.
4. Results
4.1. Extraction of Stem Point Clouds
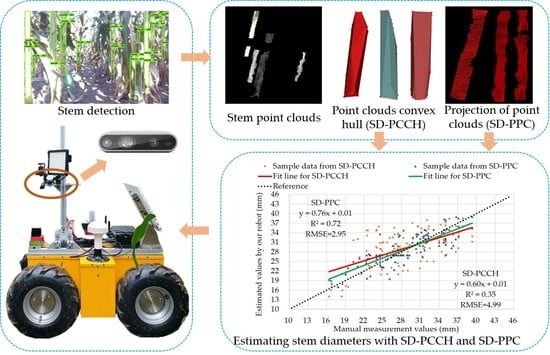
4.2. Visualization of Convex Hull and 2D Projection of Point Cloud
4.3. Point Cloud Filling
4.4. Stem Diameter Estimation with SD-PCCH and SD-PPC
5. Discussion
- (i)
- Improving the stem detection accuracy of convolutional neural network.We used the existing two-stage object detector Faster-RCNN to identify field stems. The mAP of stem detection after network convergence was 67%. This may be caused by the strong lighting changes and the inconspicuous color characteristics of stems under the crop canopy. In the future, we hope to improve the detection accuracy of the detector by labeling more data sets and adjusting the network structure;
- (ii)
- Evaluating the 3D image quality of RealSense D435i.RealSense D435i cameras have been proven to have excellent ranging performances under natural conditions. However, it is still necessary to evaluate the depth value accuracy for different crop organs to improve the 3D imaging quality. It will be helpful to improve the measurement accuracy of maize stem diameters;
- (iii)
- Improving the real-time phenotyping performances of our algorithm pipeline.Currently, our algorithm pipeline is implemented on a graphics workstation. During our experiment, the bag recording function of ROS was used to obtain crop images in the field. These image data were parsed and used on the graphics workstation to run our phenotyping algorithm. In the future, our algorithm will be processed in real-time with an edge computing module on our HTP robot;
- (iv)
- Extending our algorithm pipeline to different crop varieties.At present, maize crops are our main focus. However, the algorithm pipeline we proposed is expected to be applied to other common high-stem plants, such as sorghum, sugarcane, etc. Furthermore, we believe that our method can also be used for the measurement of the phenotypic parameters of various crop organs by only adjusting some necessary algorithm parameters.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef] [Green Version]
- Hunter, M.C.; Smith, R.G.; Schipanski, M.E.; Atwood, L.W.; Mortensen, D.A. Agriculture in 2050: Recalibrating Targets for Sustainable Intensification. BioScience 2017, 67, 386–391. [Google Scholar] [CrossRef] [Green Version]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Quan, C.; Song, Z.; Li, X.; Yu, G.; Li, C.; Muhammad, A. High-Throughput Plant Phenotyping Platform (HT3P) as a Novel Tool for Estimating Agronomic Traits from the Lab to the Field. Front. Bioeng. Biotechnol. 2020, 8, 623705. [Google Scholar] [CrossRef]
- Mir, R.R.; Reynolds, M.; Pinto, F.; Khan, M.A.; Bhat, M.A. High-throughput phenotyping for crop improvement in the genomics era. Plant Sci. 2019, 282, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Wang, J.; Guo, X.; Yang, W.; Zhao, C. High-throughput phenotyping: Breaking through the bottleneck in future crop breeding. Crop J. 2021, 9, 633–645. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Atefi, A.; Ge, Y.; Pitla, S.; Schnable, J. Robotic Technologies for High-Throughput Plant Phenotyping: Contemporary Reviews and Future Perspectives. Front. Plant Sci. 2021, 12, 611940. [Google Scholar] [CrossRef]
- Robertson, D.J.; Julias, M.; Lee, S.Y.; Cook, D.D. Maize Stalk Lodging: Morphological Determinants of Stalk Strength. Crop Sci. 2017, 57, 926–934. [Google Scholar] [CrossRef] [Green Version]
- Vit, A.; Shani, G. Comparing RGB-D Sensors for Close Range Outdoor Agricultural Phenotyping. Sensors 2018, 18, 4413. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Sun, N.; Qiu, Q.; Li, T.; Zhao, C. Depth Ranging Performance Evaluation and Improvement for RGB-D Cameras on Field-Based High-Throughput Phenotyping Robots *. In Proceedings of the 2021 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Prague, Czech Republic, 27 September–1 October 2021; pp. 3299–3304. [Google Scholar]
- Araus, J.L.; Kefauver, S.C.; Diaz, O.V.; Gracia-Romero, A.; Rezzouk, F.Z.; Segarra, J.; Buchaillot, M.L.; Chang-Espino, M.; Vatter, T.; Sanchez-Bragado, R.; et al. Crop phenotyping in a context of Global Change: What to measure and how to do it. J. Integr. Plant Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Chaudhary, V.; Yadav, R.C.; Yadav, N.R. High-Throughput Phenotyping: A Platform to Accelerate Crop Improvement. Phenomics 2021, 1, 31–53. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2017, 44, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, G.; Kim, J.; Yu, J.-K.; Kim, H.-J.; Kim, Y.; Kim, D.-W.; Kim, K.-H.; Lee, C.W.; Chung, Y.S. Review: Cost-Effective Unmanned Aerial Vehicle (UAV) Platform for Field Plant Breeding Application. Remote Sens. 2020, 12, 998. [Google Scholar] [CrossRef] [Green Version]
- Araus, J.L.; Kefauver, S.C. Breeding to adapt agriculture to climate change: Affordable phenotyping solutions. Curr. Opin. Plant Biol. 2018, 45, 237–247. [Google Scholar] [CrossRef]
- Jin, X.; Zarco-Tejada, P.J.; Schmidhalter, U.; Reynolds, M.P.; Hawkesford, M.J.; Varshney, R.K.; Yang, T.; Nie, C.; Li, Z.; Ming, B.; et al. High-Throughput Estimation of Crop Traits: A Review of Ground and Aerial Phenotyping Platforms. IEEE Geosci. Remote Sens. Mag. 2021, 9, 200–231. [Google Scholar] [CrossRef]
- Shafiekhani, A.; Kadam, S.; Fritschi, F.B.; DeSouza, G.N. Vinobot and Vinoculer: Two Robotic Platforms for High-Throughput Field Phenotyping. Sensors 2017, 17, 214. [Google Scholar] [CrossRef]
- Shafiekhani, A.; Fritschi, F.B.; DeSouza, G.N. Vinobot and vinoculer: From real to simulated platforms. In Proceedings of the SPIE Commercial + Scientific Sensing and Imaging, Orlando, FL, USA, 15–19 April 2018. [Google Scholar]
- Ahmadi, A.; Nardi, L.; Chebrolu, N.; Stachniss, C. Visual Servoing-based Navigation for Monitoring Row-Crop Fields. In Proceedings of the 2020 IEEE International Conference on Robotics and Automation (ICRA), Paris, France, 31 May–31 August 2020; pp. 4920–4926. [Google Scholar]
- Mueller-Sim, T.; Jenkins, M.; Abel, J.; Kantor, G. The Robotanist: A ground-based agricultural robot for high-throughput crop phenotyping. In Proceedings of the 2017 IEEE International Conference on Robotics and Automation (ICRA), Singapore, 29 May–3 June 2017; pp. 3634–3639. [Google Scholar]
- Kayacan, E.; Young, S.N.; Peschel, J.M.; Chowdhary, G. High-precision control of tracked field robots in the presence of unknown traction coefficients. J. Field Robot. 2018, 35, 1050–1062. [Google Scholar] [CrossRef]
- Bao, Y.; Tang, L.; Srinivasan, S.; Schnable, P.S. Field-based architectural traits characterisation of maize plant using time-of-flight 3D imaging. Biosyst. Eng. 2019, 178, 86–101. [Google Scholar] [CrossRef]
- Baharav, T.; Bariya, M.; Zakhor, A. In Situ Height and Width Estimation of Sorghum Plants from 2.5 d Infrared Images. Electron. Imaging 2017, 2017, 122–135. [Google Scholar] [CrossRef] [Green Version]
- Baweja, H.S.; Parhar, T.; Mirbod, O.; Nuske, S. StalkNet: A Deep Learning Pipeline for High-Throughput Measurement of Plant Stalk Count and Stalk Width. In Field and Service Robotics; Springer Proceedings in Advanced Robotics; Springer: Berlin/Heidelberg, Germany, 2018; pp. 271–284. [Google Scholar]
- Parhar, T.; Baweja, H.; Jenkins, M.; Kantor, G. A Deep Learning-Based Stalk Grasping Pipeline. In Proceedings of the 2018 IEEE International Conference on Robotics and Automation (ICRA), Brisbane, Australia, 21–25 May 2018; pp. 6161–6167. [Google Scholar]
- Young, S.N.; Kayacan, E.; Peschel, J.M. Design and field evaluation of a ground robot for high-throughput phenotyping of energy sorghum. Precis. Agric. 2018, 20, 697–722. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Kayacan, E.; Thompson, B.; Chowdhary, G. High precision control and deep learning-based corn stand counting algorithms for agricultural robot. Auton. Robot. 2020, 44, 1289–1302. [Google Scholar] [CrossRef]
- Qiu, Q.; Sun, N.; Bai, H.; Wang, N.; Fan, Z.; Wang, Y.; Meng, Z.; Li, B.; Cong, Y. Field-Based High-Throughput Phenotyping for Maize Plant Using 3D LiDAR Point Cloud Generated With a “Phenomobile”. Front. Plant Sci. 2019, 10, 554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Q.; Fan, Z.; Meng, Z.; Zhang, Q.; Cong, Y.; Li, B.; Wang, N.; Zhao, C. Extended Ackerman Steering Principle for the coordinated movement control of a four wheel drive agricultural mobile robot. Comput. Electron. Agric. 2018, 152, 40–50. [Google Scholar] [CrossRef]
- Qiu, R.; Wei, S.; Zhang, M.; Li, H.; Sun, H.; Liu, G.; Li, M. Sensors for measuring plant phenotyping: A review. Int. J. Agric. Biol. Eng. 2018, 11, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Su, Y.; Gao, S.; Wu, F.; Ma, Q.; Xu, K.; Ma, Q.; Hu, T.; Liu, J.; Pang, S.; et al. Separating the Structural Components of Maize for Field Phenotyping Using Terrestrial LiDAR Data and Deep Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2020, 58, 2644–2658. [Google Scholar] [CrossRef]
- Kazmi, W.; Foix, S.; Alenyà, G.; Andersen, H.J. Indoor and outdoor depth imaging of leaves with time-of-flight and stereo vision sensors: Analysis and comparison. ISPRS J. Photogramm. Remote Sens. 2014, 88, 128–146. [Google Scholar] [CrossRef] [Green Version]
- Hämmerle, M.; Höfle, B. Mobile low-cost 3D camera maize crop height measurements under field conditions. Precis. Agric. 2017, 19, 630–647. [Google Scholar] [CrossRef]
- Kurtser, P.; Ringdahl, O.; Rotstein, N.; Berenstein, R.; Edan, Y. In-Field Grape Cluster Size Assessment for Vine Yield Estimation Using a Mobile Robot and a Consumer Level RGB-D Camera. IEEE Robot. Autom. Lett. 2020, 5, 2031–2038. [Google Scholar] [CrossRef]
- Atefi, A.; Ge, Y.; Pitla, S.; Schnable, J. Robotic Detection and Grasp of Maize and Sorghum: Stem Measurement with Contact. Robotics 2020, 9, 58. [Google Scholar] [CrossRef]
- Li, Y.; Wen, W.; Guo, X.; Yu, Z.; Gu, S.; Yan, H.; Zhao, C. High-throughput phenotyping analysis of maize at the seedling stage using end-to-end segmentation network. PLoS ONE 2021, 16, e0241528. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hu, P.; Zheng, B.; Duan, T.; Zhu, B.; Guo, Y. A field-based high-throughput method for acquiring canopy architecture using unmanned aerial vehicle images. Agric. For. Meteorol. 2021, 296, 108231. [Google Scholar] [CrossRef]
- Zermas, D.; Morellas, V.; Mulla, D.; Papanikolopoulos, N. 3D model processing for high throughput phenotype extraction–the case of corn. Comput. Electron. Agric. 2020, 172, 105047. [Google Scholar] [CrossRef]
- Erndwein, L.; Cook, D.D.; Robertson, D.J.; Sparks, E.E. Field-based mechanical phenotyping of cereal crops to assess lodging resistance. Appl. Plant Sci. 2020, 8, e11382. [Google Scholar] [CrossRef]
- Fan, Z.; Sun, N.; Qiu, Q.; Li, T.; Zhao, C. A High-Throughput Phenotyping Robot for Measuring Stalk Diameters of Maize Crops. In Proceedings of the 2021 IEEE 11th Annual International Conference on CYBER Technology in Automation, Control, and Intelligent Systems (CYBER), Jiaxing, China, 27–31 July 2021; pp. 128–133. [Google Scholar]
- Mahesh Kumar, K.; Rama Mohan Reddy, A. A fast DBSCAN clustering algorithm by accelerating neighbor searching using Groups method. Pattern Recognit. 2016, 58, 39–48. [Google Scholar] [CrossRef]
- Vázquez-Arellano, M.; Reiser, D.; Paraforos, D.S.; Garrido-Izard, M.; Burce, M.E.C.; Griepentrog, H.W. 3-D reconstruction of maize plants using a time-of-flight camera. Comput. Electron. Agric. 2018, 145, 235–247. [Google Scholar] [CrossRef]

















| Specifications | Parameters | Specifications | Parameters |
|---|---|---|---|
| Size | 0.80 m × 0.45 m × 0.40 m | Mass | 40 kg |
| Operating temperature | −15–50 °C | Carrying capacity | 30 kg |
| Working time | 4 h | Voltage | 24 V |
| Climbing gradient | 25° | Maximum velocity | 0.30 m/s |
| Mobile mode | Wheeled model | Obstacle clearing capability | 0.15 m |
| Steering mode | Differential steering | Ground clearance | 0.10 m |
| Working environment | In-row | Applied coding interface | ROS, C++, Python |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Z.; Sun, N.; Qiu, Q.; Li, T.; Feng, Q.; Zhao, C. In Situ Measuring Stem Diameters of Maize Crops with a High-Throughput Phenotyping Robot. Remote Sens. 2022, 14, 1030. https://doi.org/10.3390/rs14041030
Fan Z, Sun N, Qiu Q, Li T, Feng Q, Zhao C. In Situ Measuring Stem Diameters of Maize Crops with a High-Throughput Phenotyping Robot. Remote Sensing. 2022; 14(4):1030. https://doi.org/10.3390/rs14041030
Chicago/Turabian StyleFan, Zhengqiang, Na Sun, Quan Qiu, Tao Li, Qingchun Feng, and Chunjiang Zhao. 2022. "In Situ Measuring Stem Diameters of Maize Crops with a High-Throughput Phenotyping Robot" Remote Sensing 14, no. 4: 1030. https://doi.org/10.3390/rs14041030
APA StyleFan, Z., Sun, N., Qiu, Q., Li, T., Feng, Q., & Zhao, C. (2022). In Situ Measuring Stem Diameters of Maize Crops with a High-Throughput Phenotyping Robot. Remote Sensing, 14(4), 1030. https://doi.org/10.3390/rs14041030