Estimating Crop Sowing and Harvesting Dates Using Satellite Vegetation Index: A Comparative Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
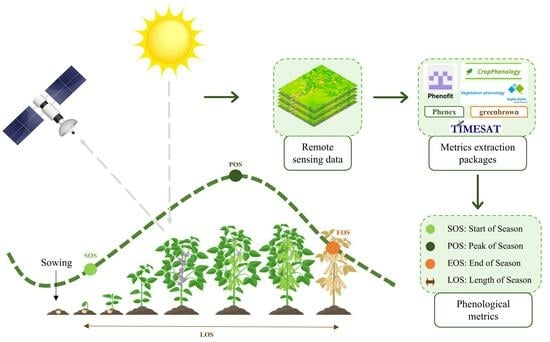
2.2. Remote Sensing Data
2.3. Satellite Data Pre-Processing
2.3.1. MOD13Q1/MYD13Q1 Processing
2.3.2. MOD09Q1 Processing
2.3.3. Sentinel-2 Processing
2.4. Sowing and Harvest Dates Estimation
2.4.1. Time-Series Smoothing
2.4.2. Phenology Extraction Algorithms
- a.
- CropPhenology
- b.
- Digital Earth Australia tools package
- c.
- Greenbrown
- d.
- Phenofit
- e.
- Phenex
- f.
- TIMESAT
2.5. Parameter Calibration and Extraction
2.6. Performance Evaluation and Spatialization
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Wang, J.; Gao, F.; Liu, Y.; Schaaf, C.; Friedl, M.; Yu, Y.; Jayavelu, S.; Gray, J.; Liu, L.; et al. Exploration of scaling effects on coarse resolution land surface phenology. Remote Sens. Environ. 2017, 190, 318–330. [Google Scholar] [CrossRef]
- Jönsson, P.; Eklundh, L. Seasonality extraction by function fitting to time-series of satellite sensor data. IEEE Trans. Geosci. Remote Sens. 2002, 40, 1824–1832. [Google Scholar] [CrossRef]
- Chen, J.; Jönsson, P.; Tamura, M.; Gu, Z.; Matsushita, B.; Eklundh, L. A simple method for reconstructing a high-quality NDVI time-series data set based on the Savitzky-Golay filter. Remote Sens. Environ. 2004, 91, 332–334. [Google Scholar] [CrossRef]
- White, M.A.; de Beurs, K.M.; Didan, K.; Inouye, D.W.; Richardson, A.D.; Jensen, O.P.; O’Keefe, J.; Zhang, G.; Nemani, R.R.; van Leeuwen, W.J.; et al. Intercomparison, interpretation, and assessment of spring phenology in North America estimated from remote sensing for 1982–2006. Glob. Chang. Biol. 2009, 15, 2335–2359. [Google Scholar] [CrossRef]
- Berra, E.F.; Gaulton, R. Remote sensing of temperate and boreal forest phenology: A review of progress, challenges and opportunities in the intercomparison of in-situ and satellite phenological metrics. For. Ecol. Manag. 2021, 480, 118663. [Google Scholar] [CrossRef]
- Bolton, D.K.; Friedl, M.A. Forecasting crop yield using remotely sensed vegetation indices and crop phenology metrics. Agric. For. Meteorol. 2013, 173, 74–84. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, L.; Liu, M.; Zhang, J.; Leng, S.; Diao, C. Establishing and assessing the Integrated Surface Drought Index (ISDI) for agricultural drought monitoring in mid-eastern China. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 397–410. [Google Scholar] [CrossRef]
- Sakamoto, T.; Gitelson AA Arkebauer, T.J. MODIS-based corn grain yield estimation model incorporating crop phenology information. Remote Sens. Environ. 2013, 131, 215–231. [Google Scholar] [CrossRef]
- Gao, F.; Anderson, M.C.; Zhang, X.; Yang, Z.; Alfieri, J.G.; Kustas, W.P.; Mueller, R.; Johnson, D.M.; Prueger, J.H. Toward mapping crop progress at field scales through fusion of Landsat and MODIS imagery. Remote Sens. Environ. 2017, 188, 9–25. [Google Scholar] [CrossRef]
- Diao, C. Remote sensing phenological monitoring framework to characterize corn and soybean physiological growing stages. Remote Sens. Environ. 2020, 248, 111960. [Google Scholar] [CrossRef]
- Zeng, L.; Wardlow, B.D.; Xiang, D.; Hu, S.; Li, D. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Jung, J.; Maeda, M.; Chang, A.; Bhandari, M.; Ashapure, A.; Landivar-Bowles, J. The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr. Opin. Biotechnol. 2021, 70, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Pan, Y.; Zhu, X.; Wang, J.; Li, Q. Prediction of crop yield using phenological information extracted from remote sensing vegetation index. Sensors 2021, 21, 1406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Abrahao, G.; Cohn, A.; Campolo, J.; Thompson, S. A MODIS-based scalable remote sensing method to estimate sowing and harvest dates of soybean crops in Mato Grosso, Brazil. Heliyon 2021, 7, e07436. [Google Scholar] [CrossRef] [PubMed]
- Belda, S.; Pipia, L.; Morcillo-Pallarés, P.; Rivera-Caicedo, J.P.; Amin, E.; Grave, C.D.; Verrelst, J. DATimeS: A machine learning time series GUI toolbox for gap-filling and vegetation phenology trends detection. Environ. Model. Softw. 2020, 127, 104666. [Google Scholar] [CrossRef]
- Sadeh, Y.; Zhu, X.; Chenu, K.; Dunkerley, D. Sowing date detection at the field scale using CubeSats remote sensing. Comput. Electron. Agric. 2019, 157, 568–580. [Google Scholar] [CrossRef]
- Araya, S.; Ostendorf, B.; Lyle, G.; Lewis, M. CropPhenology: An R package for extracting crop phenology from time series remotely sensed vegetation index imagery. Ecol. Inform. 2018, 46, 45–56. [Google Scholar] [CrossRef]
- Duarte, L.; Teodoro, A.C.; Monteiro, A.T.; Cunha, M.; Gonçalves, H. QPhenoMetrics: An open-source software application to assess vegetation phenology metrics. Comput. Electron. Agric. 2018, 148, 82–94. [Google Scholar] [CrossRef]
- Duarte, L.; Scomparim, S.; Teodoro, A.C. Vegetation phenology from Sentinel 2 data: A GIS open source application. In Proceedings of the Conference on Earth Resources and Environmental Remote Sensing/GIS Applications XIII, Berlin, Germany, 5–7 September 2022; Volume 12268, pp. 183–189. [Google Scholar] [CrossRef]
- Kong, D.; McVicar, T.R.; Xiao, M.; Zhang, Y.; Peña-Arancibia, J.L.; Filippa, G.; Xie, Y.; Gu, X. phenofit: An R package for extracting vegetation phenology from time series remote sensing. Methods Ecol. Evol. 2022, 13, 1508–1527. [Google Scholar] [CrossRef]
- Tan, B.; Morisette, J.T.; Wolfe, R.E.; Gao, F.; Ederer, G.A.; Nightingale, J.; Pedelty, J.A. An enhanced TIMESAT algorithm for estimating vegetation phenology metrics from MODIS data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 361–371. [Google Scholar] [CrossRef]
- Palmer, S.C.; Odermatt, D.; Hunter, P.D.; Brockmann, C.; Presing, M.; Balzter, H.; Tóth, V.R. Satellite remote sensing of phytoplankton phenology in Lake Balaton using 10 years of MERIS observations. Remote Sens. Environ. 2015, 158, 441–452. [Google Scholar] [CrossRef]
- Lange, M.; Doktor, D. phenex: Auxiliary Functions for Phenological Data Analysis, R Package Version 1.4-5. 2017. Available online: https://cran.r-project.org/web/packages/phenex/phenex.pdf (accessed on 5 November 2022).
- Forkel, M.; Wutzler, T. Greenbrown—Land Surface Phenology and Trend Analysis. A Package for the R Software. Version 2.2. 15 April 2015. Available online: http://greenbrown.r-forge.r-project.org/ (accessed on 1 March 2023).
- Kong, D. rTIMESAT: Extract Remote Sensing Vegetation Phenology by TIMESAT Fortran Library. 2021. Available online: https://rdrr.io/github/kongdd/rTIMESAT/ (accessed on 20 August 2022).
- Digital Earth Australia—DEA. Vegetation Phenology. Available online: https://docs.dea.ga.gov.au/notebooks/Real_world_examples/Vegetation_phenology.html (accessed on 4 December 2022).
- Misra, G.; Buras, A.; Menzel, A. Effects of different methods on the comparison between land surface and ground phenology—A methodological case study from south-western Germany. Remote Sens. 2016, 8, 753. [Google Scholar] [CrossRef]
- D’Odorico, P.; Gonsamo, A.; Gough, C.M.; Bohrer, G.; Morison, J.; Wilkinson, M.; Hanson, P.J.; Gianelle, D.; Fuentes, J.D.; Buchmann, N. The match and mismatch between photosynthesis and land surface phenology of deciduous forests. Agric. For. Meteorol. 2015, 214, 25–38. [Google Scholar] [CrossRef]
- Mariethoz, G.; McCabe, M.F.; Renard, P. Spatiotemporal reconstruction of gaps in multivariate fields using the direct sampling approach. Water Resour. Res. 2012, 48, 1–13. [Google Scholar] [CrossRef]
- Ahl, D.E.; Gower, S.T.; Burrows, S.N.; Shabanov, N.V.; Myneni, R.B.; Knyazikhin, Y. Monitoring spring canopy phenology of a deciduous broadleaf forest using MODIS. Remote Sens. Environ. 2006, 104, 88–95. [Google Scholar] [CrossRef]
- Siłuch, M.; Bartmiński, P.; Zgłobicki, W. Remote Sensing in Studies of the Growing Season: A Bibliometric Analysis. Remote Sens. 2022, 14, 1331. [Google Scholar] [CrossRef]
- Chen, F.; Liu, Z.; Zhong, H.; Wang, S. Exploring the applicability and scaling effects of satellite-observed spring and autumn phenology in complex terrain regions using four different spatial resolution products. Remote Sens. 2021, 13, 4582. [Google Scholar] [CrossRef]
- Liu, L.; Cao, R.; Shen, M.; Chen, J.; Wang, J.; Zhang, X. How does scale effect influence spring vegetation phenology estimated from satellite-derived vegetation indexes? Remote Sens. 2019, 11, 2137. [Google Scholar] [CrossRef]
- Fisher, J.I.; Mustard, J.F. Cross-scalar satellite phenology from ground, Landsat, and MODIS data. Remote Sens. Environ. 2007, 109, 261–273. [Google Scholar] [CrossRef]
- Rezaei, E.E.; Ghazaryan, G.; González, J.; Cornish, N.; Dubovyk, O.; Siebert, S. The use of remote sensing to derive maize sowing dates for large-scale crop yield simulations. Int. J. Biometeorol. 2021, 65, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.R.; Silva LC, D.A.; Richetti, J.; Ló, T.B.; Johann, J.A. Harvest date forecast for soybeans from maximum vegetative development using satellite images. Int. J. Remote Sens. 2021, 42, 1121–1138. [Google Scholar] [CrossRef]
- Helman, D. Land surface phenology: What do we really ‘see’ from space? Sci. Total Environ. 2018, 618, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Song, X.P.; Hansen, M.C.; Potapov, P.; Adusei, B.; Pickering, J.; Adami, M.; Lima, A.; Zalles, V.; Stehman, S.V.; Di Bella, C.M.; et al. Massive soybean expansion in South America since 2000 and implications for conservation. Nat. Sustain. 2021, 4, 784–792. [Google Scholar] [CrossRef]
- Johann, J.A.; Becker, W.R.; Uribe-Opazo, M.A.; Mercante, E. Uso de imagens do sensor orbital MODIS na estimação de datas do ciclo de desenvolvimento da cultura da soja para o estado do Paraná—Brasil. Eng. Agríc. 2016, 36, 126–142. [Google Scholar] [CrossRef]
- Becker, W.R.; Richetti, J.; Mercante, E.; Júlio Dalla, C.; Esquerdo, M.; Carlos Da, A.; Junior, S.; Paludo, A.; Johann, J.A. Agricultural soybean and corn calendar based on moderate resolution satellite images for southern Brazil Calendário agrícola de soja e milho baseado em imagens de satélite de moderada resolução para o sul do Brasil. Ciênc. Agrár. 2020, 41, 2419–2428. [Google Scholar] [CrossRef]
- CONAB—Companhia Nacional de Abastecimento. Calendário de Plantio e Colheita de Grãos no Brasil. 2022. Available online: https://www.conab.gov.br/institucional/publicacoes/outras-publicacoes (accessed on 6 August 2023).
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Huang, X.; Liu, J.; Zhu, W.; Atzberger, C.; Liu, Q. The optimal threshold and vegetation index time series for retrieving crop phenology based on a modified dynamic threshold method. Remote Sens. 2019, 11, 2725. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Townshend, J.R.G.; Huang, C.; Kalluri, S.N.V.; Defries, R.S.; Liang, S.; Yang, K. Beware of per-pixel characterization of land cover. Int. J. Remote Sens. 2000, 21, 839–843. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Viovy, N.; Arino, O.; Belward, A.S. The Best Index Slope Extraction (BISE): A method for reducing noise in NDVI time-series. Int. J. Remote Sens. 1992, 13, 1585–1590. [Google Scholar] [CrossRef]
- Dhu, T.; Dunn, B.; Lewis, B.; Lymburner, L.; Mueller, N.; Telfer, E.; Lewis, A.; McIntyre, A.; Minchin, S.; Phillips, C. Digital earth Australia–unlocking new value from earth observation data. Big Earth Data 2017, 22, 64–74. [Google Scholar] [CrossRef]
- Forkel, M.; Migliavacca, M.; Thonicke, K.; Reichstein, M.; Schaphoff, S.; Weber, U.; Carvalhais, N. Codominant water control on global interannual variability and trends in land surface phenology and greenness. Glob. Chang. Biol. 2015, 21, 3414–3435. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, P.; Eklundh, L. TIMESAT—A program for analysing time-series of satellite sensor data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Geng, L.; Ma, M.; Wang, X.; Yu, W.; Jia, S.; Wang, H. Comparison of eight techniques for reconstructing multi-satellite sensor time-series NDVI data sets in the Heihe river basin, China. Remote Sens. 2014, 6, 2024–2049. [Google Scholar] [CrossRef]
- Urban, D.; Guan, K.; Jain, M. Estimating sowing dates from satellite data over the US Midwest: A comparison of multiple sensors and metrics. Remote Sens. Environ. 2018, 211, 400–412. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, Y.; Qin, Y.; Xia, H.; Lu, H.; Liu, S.; Li, N.; Fu, Y. Evaluating the accuracy of and evaluating the potential errors in extracting vegetation phenology through remote sensing in China. Int. J. Remote Sens. 2020, 41, 3592–3613. [Google Scholar] [CrossRef]
- Trentin, R.; Heldwein, A.B.; Streck, N.A.; Trentin, G.; Silva, J.C.D. Subperíodos fenológicos e ciclo da soja conforme grupos de maturidade e datas de semeadura. Pesqui. Agropecu. Bras. 2013, 48, 703–713. [Google Scholar] [CrossRef]
- Zanon, A.J.; Winck, J.E.; Streck, N.A.; Rocha, T.S.; Cera, J.C.; Richter, G.L.; Lago, I.; Santos, P.M.; Maciel, L.D.; Guedes, J.V.; et al. Desenvolvimento de cultivares de soja em função do grupo de maturação e tipo de crescimento em terras altas e terras baixas. Bragantia 2015, 21, 400–411. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Sidney, S. Nonparametric statistics for the behavioral sciences. J. Nerv. Ment. Dis. 1957, 125, 497. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, C.; Liu, Y.; Wang, X.; Fang, B.; Yuan, W.; Ge, Q. Spring green-up date derived from GIMMS3g and SPOT-VGT NDVI of winter wheat cropland in the North China Plain. ISPRS J. Photogramm. Remote Sens. 2017, 130, 81–91. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, E.; Kirkegaard, J.A.; Rebetzke, G.J. Novel wheat varieties facilitate deep sowing to beat the heat of changing climates. Nat. Clim. Chang. 2022, 12, 291–296. [Google Scholar] [CrossRef]
- Lawes, R.; Mata, G.; Richetti, J.; Fletcher, A.; Herrmann, C. Using remote sensing, process-based crop models, and machine learning to evaluate crop rotations across 20 million hectares in Western Australia. Agron. Sustain. Dev. 2022, 42, 120. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B. Sensitivity of vegetation phenology detection to the temporal resolution of satellite data. Int. J. Remote Sens. 2009, 30, 2061–2074. [Google Scholar] [CrossRef]
- Garrity, S.R.; Bohrer, G.; Maurer, K.D.; Mueller, K.L.; Vogel, C.S.; Curtis, P.S. A comparison of multiple phenology data sources for estimating seasonal transitions in deciduous forest carbon exchange. Agric. For. Meteorol. 2011, 151, 1741–1752. [Google Scholar] [CrossRef]
- Guyon, D.; Guillot, M.; Vitasse, Y.; Cardot, H.; Hagolle, O.; Delzon, S.; Wigneron, J.P. Monitoring elevation variations in leaf phenology of deciduous broadleaf forests from SPOT/VEGETATION time-series. Remote Sens. Environ. 2011, 115, 615–627. [Google Scholar] [CrossRef]
- Wu, C.; Gonsamo, A.; Gough, C.M.; Chen, J.M.; Xu, S. Modeling growing season phenology in North American forests using seasonal mean vegetation indices from MODIS. Remote Sens. Environ. 2014, 147, 79–88. [Google Scholar] [CrossRef]
- Rao, N.R.; Garg, P.K.; Ghosh, S.K. Development of an agricultural crops spectral library and classification of crops at cultivar level using hyperspectral data. Precis. Agric. 2007, 8, 173–185. [Google Scholar] [CrossRef]
- Wang, S.; Mo, X.; Liu, Z.; Baig MH, A.; Chi, W. Understanding long-term (1982–2013) patterns and trends in winter wheat spring green-up date over the North China Plain. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 235–244. [Google Scholar] [CrossRef]
- Bendini, H.D.N.; Fonseca, L.M.; Schwieder, M.; Körting, T.S.; Rufin, P.; Sanches, I.D.; Leitão, P.J.; Hostert, P. Detailed agricultural land classification in the Brazilian cerrado based on phenological information from dense satellite image time series. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101872. [Google Scholar] [CrossRef]
- Matongera, T.N.; Mutanga, O.; Sibanda, M.; Odindi, J. Estimating and monitoring land surface phenology in rangelands: A review of progress and challenges. Remote Sens. 2021, 13, 2060. [Google Scholar] [CrossRef]
- Manfron, G.; Delmotte, S.; Busetto, L.; Hossard, L.; Ranghetti, L.; Brivio, P.A.; Boschetti, M. Estimating inter-annual variability in winter wheat sowing dates from satellite time series in Camargue, France. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 190–201. [Google Scholar] [CrossRef]
- Lobell, D.B.; Ortiz-Monasterio, J.I.; Sibley, A.M.; Sohu, V.S. Satellite detection of earlier wheat sowing in India and implications for yield trends. Agric. Syst. 2013, 115, 137–143. [Google Scholar] [CrossRef]
- Vyas, S.; Nigam, R.; Patel, N.K.; Panigrahy, S. Extracting regional pattern of wheat sowing dates using multispectral and high temporal observations from Indian geostationary satellite. J. Indian Soc. Remote Sens. 2013, 41, 855–864. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, Y.; Ren, G.; Wang, G. Precipitation trends over mainland China from 1961–2016 after removal of measurement biases. J. Geophys. Res. Atmos. 2020, 125, e2019JD031728. [Google Scholar] [CrossRef]
- Araza, A.; de Bruin, S.; Herold, M.; Quegan, S.; Labriere, N.; Rodriguez-Veiga, P.; Avitabile, V.; Santoro, M.; Mitchard, E.T.; Ryan, C.M.; et al. A comprehensive framework for assessing the accuracy and uncertainty of global above-ground biomass maps. Remote Sens. Environ. 2022, 272, 112917. [Google Scholar] [CrossRef]
- McNairn, H.; Jiao, X.; Pacheco, A.; Sinha, A.; Tan, W.; Li, Y. Estimating canola phenology using synthetic aperture radar. Remote Sens. Environ. 2018, 219, 196–205. [Google Scholar] [CrossRef]
- Claverie, M.; Ju, J.; Masek, J.G.; Dungan, J.L.; Vermote, E.F.; Roger, J.C.; Skakun, S.V.; Justice, C. The Harmonized Landsat and Sentinel-2 surface reflectance data set. Remote Sens. Environ. 2018, 219, 145–161. [Google Scholar] [CrossRef]
- Houborg, R.; McCabe, M.F. Daily Retrieval of NDVI and LAI at 3 m Resolution via the Fusion of CubeSat, Landsat, and MODIS Data. Remote Sens. 2018, 10, 890. [Google Scholar] [CrossRef]
- Houborg, R.; McCabe, M.F. A cubesat enabled spatio-temporal enhancement method (cestem) utilizing planet, landsat and modis data. Remote Sens. Environ. 2018, 209, 211–226. [Google Scholar] [CrossRef]
- Younes, N.; Joyce KE Maier, S.W. All models of satellite-derived phenology are wrong, but some are useful: A case study from northern Australia. Int. J. Appl. Earth Obs. Geoinf. 2021, 97, 102285. [Google Scholar] [CrossRef]








| Package (Parameters) | CP (Threshold) | DT (Derivative) | PX (Threshold) | TM (Threshold) |
|---|---|---|---|---|
| MCD13 | (0.25, 0.35) | (median, last) | (0.15, 0.15) | (0.15, 0.20) |
| MOD09 | (0.30, 0.35) | (median, last) | (0.15, 0.15) | (0.15, 0.20) |
| MOD13 | (0.25, 0.25) | (median, last) | (0.15, 0.15) | (0.15, 0.20) |
| Sentinel-2 | (0.10, 0.40) | (median, last) | (0.10, 0.10) | (0.10, 0.10) |
| CP | DT | GB | PF | PX | TM | |
|---|---|---|---|---|---|---|
| Time (s) | 0.343 | 6.879 | 0.130 | 0.146 | 10.278 | 76.544 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodigheri, G.; Sanches, I.D.; Richetti, J.; Tsukahara, R.Y.; Lawes, R.; Bendini, H.d.N.; Adami, M. Estimating Crop Sowing and Harvesting Dates Using Satellite Vegetation Index: A Comparative Analysis. Remote Sens. 2023, 15, 5366. https://doi.org/10.3390/rs15225366
Rodigheri G, Sanches ID, Richetti J, Tsukahara RY, Lawes R, Bendini HdN, Adami M. Estimating Crop Sowing and Harvesting Dates Using Satellite Vegetation Index: A Comparative Analysis. Remote Sensing. 2023; 15(22):5366. https://doi.org/10.3390/rs15225366
Chicago/Turabian StyleRodigheri, Grazieli, Ieda Del’Arco Sanches, Jonathan Richetti, Rodrigo Yoiti Tsukahara, Roger Lawes, Hugo do Nascimento Bendini, and Marcos Adami. 2023. "Estimating Crop Sowing and Harvesting Dates Using Satellite Vegetation Index: A Comparative Analysis" Remote Sensing 15, no. 22: 5366. https://doi.org/10.3390/rs15225366
APA StyleRodigheri, G., Sanches, I. D., Richetti, J., Tsukahara, R. Y., Lawes, R., Bendini, H. d. N., & Adami, M. (2023). Estimating Crop Sowing and Harvesting Dates Using Satellite Vegetation Index: A Comparative Analysis. Remote Sensing, 15(22), 5366. https://doi.org/10.3390/rs15225366