The overall aim of this study was to evaluate whether the fusion of PH and VIs can improve the predictability of dry and fresh barley biomass compared to each parameter as individual estimator. For this work, the use of TLS to derive PH was verified and bivariate BRMs based on PH or one of six VIs as well as multivariate BRMs based on the fusion of PH with each VI were established. Extensive fieldwork over three years supported the practical application of the presented methods for monitoring crop development on plot level. The same instruments were used for all measurements whereby variations through different sensors could be excluded. However, the design of the field experiment and the measurement program was slightly modified and optimized over the years. Hence, only a part of the acquired data was used for the final model generation in order to ensure the comparability between the data sets. In the following, first the retrieval of PH from TLS data is discussed before the different BRMs are examined.
4.1. TLS-Derived Plant Height
The presented study verified the reliability of the laser scanner Riegl LMS-Z420i for capturing crop surfaces. In comparison with past studies [
31,
43], the scanning angle to the field was optimized through the elevated position on the hydraulic platform. However, uncertainties still remain about the influence of the scanning angle and the fixed position of the scanner during the measurements. As maintained by Ehlert and Heisig [
62]—the scanning angle can cause overestimations in the height of reflection points and should be considered in the calculation of heights. In this study, the crop surface was determined from the merged and cleaned point clouds of four scan positions, filtered with a scheme for selecting maximum points. Overestimations should therefore be precluded.
For the practical implementation of CSM-derived plant height measurements, further aspects have to be considered. Usually, the factors time and cost have a major influence on choosing a system. As shown by Hämmerle and Höfle [
63] the appropriate point density for generating a CSM varies depending on the application. In further studies, cost-efficient systems, such as the Velodyne HDL-64E [
64], should be considered to investigate their potential for capturing crop surfaces in an adequate resolution. In the distant future, low-cost stationary systems might get permanently established for monitoring plant growth on field level. Moreover, recent developments have brought up new laser scanning platforms that might accelerate the field measurement process and optimize the scanning angle. First, ground-based mobile laser scanning (MLS) systems [
65] should be taken into account for increasing the homogeneous distribution in the point cloud and thus enhancing a uniform field coverage. Second, unmanned aerial vehicles (UAVs), such as the recently introduced Riegl RiCOPTER [
66], should be examined as a potential platform of a light-weight airborne laser scanning (ALS) systems. Promising results have already been achieved for measuring tree heights [
67] or detecting pruning of individual stems [
68] with UAV-based laser scanning. However, as examined in a comparative study for TLS and common plane-based ALS, the scanning angle and possible resolution influence the results [
69] and thus have also to be taken into account for studies on UAV-based scanning systems.
In this study, TLS measurements were used to derive 3D information of points. As shown in other studies, captured intensity values could be used for qualitative analyses of the points, such as detecting single plants [
70,
71]. Whilst such analyses were not an object of this study they should be considered for further investigations. Moreover, full waveform analysis, commonly known from ALS, can simplify the distinction between laser returns on canopy and ground returns in TLS data [
72,
73]. The scanner used in this study however is not capable of capturing the full waveform.
The maps of plant height demonstrate the potential of the present approach for deriving plant height information on plot level in a very high resolution. The methodology of spatial plant height mapping can be scaled to field level, as long as the maximum range of the scanner is regarded and the point density is above the required minimum. As shown by Hämmerle and Höfle [
63], the coverage of the field and attained mean heights are influenced by the point density. The approach of pixel-wise calculating plant height from TLS-derived CSMs has already shown good results at the field level for monitoring a maize field, about 80 m by 160 m in size [
74] and a sugar beet field, about 300 m by 500 m in size [
43] captured from four and eight scan positions, respectively. Further studies are necessary for determining crop- or case-specific minimum values for the point density. In this context, the used sensor and its maximum range influence the required number of scan positions.
Nevertheless, for this study, high coefficients of determination between averaged CSM-derived and manual measured plant heights validate the TLS measurements. For the absolute values, differences between the measurement methods have to be considered. Whereas for the manual measurement the heights of ten plants were averaged per plot, the CSM captured the entire crop surface. Consequently, differences in the mean heights occurred, which make precision analysis between TLS data and manual measurements infeasible. The precision of TLS measurements for agricultural applications is presumed from other studies [
26,
71]. It is important to note that a key advantage of the TLS data is that while plants for the manual measurements are subjectively selected, CSMs enable an objective assessment of spatially continuous plant height.
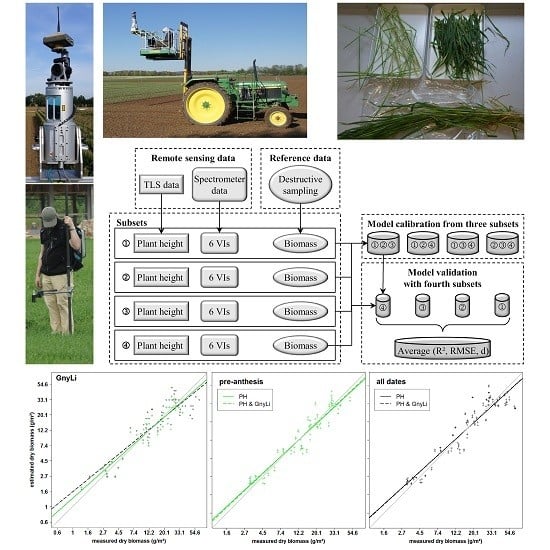
4.3. Biomass Estimation from Vegetation Indices
All VIs in this study have previously shown a relationship with biomass and LAI. Since the VIs use different bands within the spectral range, they were subdivided into three categories VIS VIs (RGBVI), VISNIR VIs (NDVI, RDVI, REIP), and NIR VIs (NRI, GnyLi). The VIs showed varying performances for the estimation of dry and fresh biomass, also depending on the regarded time frame of the growing season. Generally, the VIs within a category showed a similar behavior.
The saturation problem of the NDVI type VISNIR VIs was confirmed: Typically, crops reach 100% canopy cover around mid-vegetative phase. However, most crops continue to accumulate biomass and LAI afterwards. At a LAI of about 2.5–3, the absorbed amount of red light reaches a peak while the NIR scattering by leaves continues to increase. Thus, the ratio of NDVI type VISNIR VIs will only show slight changes [
13]. In this study, the sensitivity thresholds were about 7.4 g/m² and 55 g/m² for dry and fresh biomass, respectively. Additionally, after heading the canopy de-greens due to flowering and fruit development (after BBCH 5,
Table 2). This leads to an increased reflectance in the red part of the spectrum and thus, decreases values of the VISNIR VIs, while the biomass does not decrease. Herein, this discrepancy resulted in an inadequate model parameterization for the BRMs of the VISNIR VIs and poorer results for the whole observed period than for pre-anthesis.
A similar behavior was observable for the RGBVI. The inferior results might be explained by the fact that this VI does not take the reflectance in the NIR region into account, where most of the absorption features for biomass-related plant compounds are situated [
75]. These results align well with the ones presented by Bendig
et al. [
54], where low correlations were found for the RGBVI with biomass after booting stage (BBCH 4,
Table 2). In pre-anthesis, relationships of the RGBVI with dry and fresh biomass were similar. These results suggest that the RGBVI is mostly related with vegetation cover and not directly with biomass.
In contrast, NIRVIs, such as GnyLi and NRI, use bands only in the NIR and are thus not affected by the absorption in the red part of the spectrum, which could explain the overall more consistent and better performance of the NIR VIs, particularly after anthesis. A later saturation of these VIs aligns well with other studies [
53,
56]. Similarly, the REIP did not show any saturation effects in the pre-anthesis and yielded very good results for dry and fresh biomass. These findings can be explained by the major influence of the NIR bands that are not normalized as they are in the NDVI type VIs. Thus, the REIP saturated later than the VISNIR and VIS VIs. Nevertheless, across the whole observed period, the performance of the REIP also decreased due to saturation. The importance of the NIR domain for biomass estimation aligns with other studies [
15,
16,
53,
56] and should be further investigated. Similar to PH, the NIR VIs performed better for dry than for fresh biomass while the VISNIR VIs generally performed better with fresh biomass. This suggests that the VISNIR VIs respond more to the canopy water content and the related reflectance change in the NIR shoulder rather than directly to the biomass.
Overall, the results show that the NIR VIs perform best in the prediction of fresh and dry biomass. Moreover, the results indicate that the VIS and VISNIR VIs might not be directly related to biomass. However, no rigorous sensitivity analysis was carried out in this study but, as indicated by the results, such analyses should be carried out in the future.
In general, hyperspectral field measurements have been shown to be useful in earlier studies to estimate biomass [
14,
15,
16,
17]. However, VIs are prone to errors by illumination changes [
76] and multiangular reflection effects [
77]. So far, the influence of these effects on the estimation of plant parameters have not been comprehensively investigated and should be examined for evaluating the potential of VIs for plant parameter estimations. Moreover, ground-based spectrometer measurements are laborious and time-consuming. Automated platforms are under development in different fields of remote sensing to overcome this difficulty but they have not yet become standard. Kicherer
et al. [
78] developed a robotic platform for phenotyping grapevine based on automatic image acquisition. Results of a mobile multi-sensor phenotyping platform for phenotyping of winter wheat are presented by Kipp
et al. [
79]. Moreover, hyperspectral UAV-based systems showed promise [
55,
80,
81,
82,
83]. Unfortunately, the promising NIR domain is currently not well covered by UAV sensing systems.
4.4. Biomass Estimation with Fused Models
Leaves make up a major part of the biomass, and VIs related to biomass are often also responsive to LAI [
13,
84]. Thus, it was assumed that the spectral information would complement the PH information by adding information about the canopy density and cover.
As described above, PH and VIs showed varying performances in the estimation of fresh and dry biomass and for pre-anthesis or the whole observed period. For dry biomass in pre-anthesis, the NIR VIs performed slightly better than PH. Here, the fusion with all VIs improved the predictability, whereby the NIR and VISNIR VIs yielded the best results. This can be explained by the sensitivity of the VIs to the vegetation cover in early growth stages. For the whole observed period, PH clearly outperformed the VIs in the multivariate BRMs and only the fusion with the NIR VIs increased the predictability slightly compared to PH alone. For the VIS and VISNIR VIs, the above described saturation effects might have counteracted the positive effect of the vegetation cover estimation in the early growth stages. Additionally, for pre-anthesis and across the whole observed period, the multivariate BRMs performed similarly regardless which VI was used. This indicates that most of the prediction power can be accounted to PH.
For fresh biomass across the whole observed period, the NIR VIs performed best, followed by PH. Although the VISNIR VIs did not perform well in the bivariate BRMs, they could improve the results when fused with PH. As described above, VISNIR VIs respond to the water content. Thus, they might have complement the PH information for the estimation of fresh biomass. Still, only a slight improvement was achieved with the fused models compared to the NIR VIs alone and overall, the results of multivariate BRMs with different VIs differed only slightly.
In pre-anthesis, only the NDVI and RGBVI performed poorer than PH while the REIP performed best for the fresh biomass. In combination with PH, the results of the NDVI and RDVI were improved the most, while the latter one also achieved the best results of all fused models. For the NIR VIs and REIP none or only very minor improvements were achieved and as for the whole observed period, the water was important because it influences the reflectance in the NIR. Additionally, the VIs correspond to vegetation cover in the early growth stages. Thus, in pre-anthesis already the VIs performed well and PH only rarely contributed to the prediction power. Only the RGBVI, NDVI, and RDVI might have carried complementary information to the PH.
In this study, the NIR VIs showed the overall best performance of the VIs and seemed to carry similar information as PH. Overall, PH and NIR VIs showed the best potential for biomass estimation as individual and fused estimators. This aligns with a recent study by Marshall and Thenkabail [
15], in which they have shown the importance of PH and the NIR domain for fresh biomass estimations. The VISNIR VIs seemed to be influenced by the water content and their performance strongly depended on the regarded time frame of the growing season. Although, no comprehensive sensitivity analysis was carried out, these findings align well with other studies [
56,
85]. Further studies are needed to investigate the influence of the growing stage on the estimation, and whether estimators, which have been found as suitable in across growth stage estimations, are suitable for estimation at individual growth stages. Such in-season estimations are particularly important for applications in precision agriculture. Additionally, in this study VIs known for estimating biomass from hyperspectral data were used. Thus, the full potential of the fusion of 3D spatial and spectral data may not have been explored. Future studies should investigate whether other parts of the spectral range complement PH information better.
Overall, this study demonstrated the strength of bivariate BRMs based on PH and NIR VIs for estimating biomass, with only slight improvements achievable through multivariate models. In contrast, the weak performances of the VIS and VISNIR VIs as individual estimators were compensated through the fusion with PH. However, statements have to be limited, since the models indicated that PH contributed the most to the prediction power. In this context, it has to be noted that neither linear nor exponential models reflected the relation between estimators and biomass perfectly and thus more complex functions have to be considered, which might take the benefits of VIs, like sensitivity to water content, better in to account.
For practical applications the benefit of the fused models might be outweighed by the expenses to deploy two different systems. Referring to this, limitations through the attainable spatial and temporal resolution of each system have to be regarded. As already mentioned, TLS measurements can be scaled up to larger fields, as long as a sufficiently point density can be achieved, which has to be determined crop- and case-specific in further studies. Apart from that, laser scanning appeared as powerful tool for the non-destructive and objective assessment of spatially resolved plant height data. Statements about the accuracy of the measured plant heights are hardly possible due to the already mentioned different spatial resolution of the plant height measurements, however the averaged difference of 0.05 m between TLS-derived and manual measured plant heights corroborate the results (
Table A1). A main benefit of the field spectrometer measurements is the high credibility of the acquired spectral data, based on a large number of former studies, however the dependence on solar radiation and the small numbers of measurements per regarded spatial area, herein per plot, are the main disadvantages. Consequently, systems are required which are capable to assess larger areas in less time with the same accuracy of the results. Ideally, spatial and spectral information should be acquired directly through one sensor. For example, recently developed sensing systems and techniques allow to create hyperspectral point clouds [
86] and hyperspectral digital surface models [
83] with only one sensor and thus, derive 3D spatial and hyperspectral information at the same time. Thus, it can be expected that 3D hyperspectral information will become increasingly available and combined analysis approaches should be further developed.