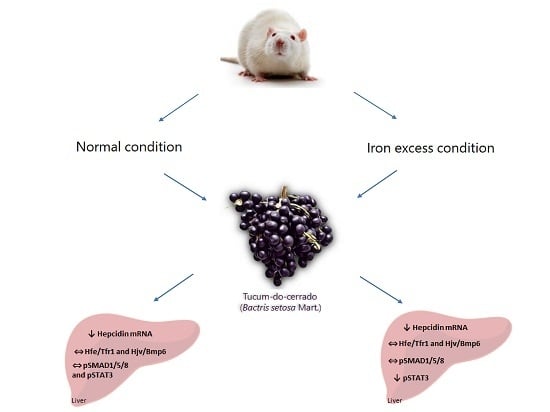
The Action of JAK/STAT3 and BMP/HJV/SMAD Signaling Pathways on Hepcidin Suppression by Tucum-do-Cerrado in a Normal and Iron-Enriched Diets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tucum-do-Cerrado Fruit Collection and Diet Preparation
2.2. Animals
2.3. Determination of the Relative Amount of Labile Iron by Electron Paramagnetic Resonance (EPR)
2.4. Determination of the mRNA Levels
2.5. Immunoblot Analysis
2.6. Statistical Analysis
3. Results
3.1. Effect of Tucum-do-Cerrado Consumption (Tuc) on the mRNA Levels of Hepatic Hepcidin (Hamp) and on the Gut Divalent Metal Transporter 1 Iron Responsive Element (Dmt1 +IRE), Divalent Metal Transporter 1 Non-Iron Responsive Element (Dmt1 -IRE), and Ferroportin (Fpn)
3.2. Effect of Tuc on the BMP6/HJV/pSMAD 1/5/8 Pathway
3.3. The Effect of Tuc on the Tfr1/Hfe Pathway
3.4. The Effect of Tuc on the Hepatic pSTAT3 Protein Levels
3.5. The Effect of Tuc on the Labile Iron Pool of the Liver, Spleen, and Intestine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Anderson, G.J.; Frazer, D.M.; McLaren, G.D. Iron absorption and metabolism. Curr. Opin. Gastroenterol. 2009, 25, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Nemeth, E. Hepcidin and disorders of iron metabolism. Annu. Rev. Med. 2011, 62, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Muckenthaler, M.U.; Galy, B.; Camaschella, C. Two to tango: Regulation of Mammalian iron metabolism. Cell 2010, 142, 24–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolas, G.; Chauvet, C.; Viatte, L.; Danan, J.L.; Bigard, X.; Devaux, I.; Beaumont, C.; Kahn, A.; Vaulont, S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Investig. 2002, 110, 1037–1044. [Google Scholar] [CrossRef]
- Sangkhae, V.; Nemeth, E. Regulation of the iron homeostatic hormone hepcidin. Adv. Nutr. 2017, 8, 126–136. [Google Scholar] [CrossRef]
- Sharp, P.A.; Clarkson, R.; Hussain, A.; Weeks, R.J.; Morison, I.M. DNA methylation of hepatic iron sensing genes and the regulation of hepcidin expression. PLoS ONE 2018, 13, e0197863. [Google Scholar] [CrossRef] [Green Version]
- Pasricha, S.R.; Lim, P.J.; Duarte, T.L.; Casu, C.; Oosterhuis, D.; Mleczko-Sanecka, K.; Suciu, M.; Da Silva, A.R.; Al-Hourani, K.; Arezes, J.; et al. Hepcidin is regulated by promoter-associated histone acetylation and HDAC3. Nat. Commun. 2017, 8, 403. [Google Scholar] [CrossRef]
- Xin, H.; Wang, M.; Tang, W.; Shen, Z.; Miao, L.; Wu, W.; Li, C.; Wang, X.; Xin, X.; Zhu, Y.Z. Hydrogen sulfide attenuates inflammatory hepcidin by reducing IL-6 secretion and promoting SIRT1-mediated STAT3 deacetylation. Antioxid. Redox Signal. 2016, 24, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Prentice, A.M. Clinical implications of new insights into hepcidin-mediated regulation of iron absorption and metabolism. Ann. Nutr. Metab. 2017, 71, 40–48. [Google Scholar] [CrossRef] [Green Version]
- Fustinoni-Reis, A.M.; Arruda, S.F.; Dourado, L.P.; da Cunha, M.S.B.; Siqueira, E.M. Tucum-Do-Cerrado (Bactris setosa Mart.) Consumption modulates iron homeostasis and prevents iron-induced oxidative stress in the rat liver. Nutrients 2016, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Rosa, F.R.; Arruda, A.F.; Siqueira, E.M.; Arruda, S.F. Phytochemical compounds and antioxidant capacity of Tucum-Do-Cerrado (Bactris setosa Mart), Brazil’s native fruit. Nutrients 2016, 8, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siqueira, E.M.; Rosa, F.R.; Fustinoni, A.M.; de Sant’Ana, L.P.; Arruda, S.F. Brazilian savanna fruits contain higher bioactive compounds content and higher antioxidant activity relative to the conventional red delicious apple. PLoS ONE 2013, 8, e72826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Cunha, M.S.B.; Arruda, S.F. Tucum-do-Cerrado (Bactris setosa Mart.) may promote anti-aging effect by upregulating SIRT1-Nrf2 pathway and attenuating oxidative stress and inflammation. Nutrients 2017, 9, 1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C., Jr. AIN-93 purified diets for laboratory rodents: Final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [CrossRef] [PubMed]
- Woodmansee, A.N.; Imlay, J.A. Quantitation of intracellular free iron by electron paramagnetic resonance spectroscopy. Methods Enzymol. 2002, 349, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, I.; Galleano, M.; Puntarulo, S. Fe allocation in liver during early stages of endotoxemia in Fe-overload rats. Toxicol. Pathol. 2011, 39, 1075–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moser, J.C.; Rawal, M.; Wagner, B.A.; Du, J.; Cullen, J.J.; Buettner, G.R. Pharmacological ascorbate and ionizing radiation (IR) increase labile iron in pancreatic cancer. Redox Biol. 2013, 2, 22–27. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, M.O.; Felipe, M.S.S.; Brígido, M.M.; Maranhão, A.Q.; De-Souza, M.T. Técnicas Básicas em Biologia Molecular, 1st ed.; Editora Universidade de Brasília: Brasília, Brazil, 2010; pp. 1–212. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hartree, E.F. Determination of protein: A modification of the Lowry method that gives a linear photometric response. Anal. Biochem. 1972, 48, 422–427. [Google Scholar] [CrossRef]
- Zhang, A.S.; Gao, J.; Koeberl, D.D.; Enns, C.A. The role of hepatocyte hemojuvelin in the regulation of bone morphogenic protein-6 and hepcidin expression in vivo. J. Biol. Chem. 2010, 285, 16416–16423. [Google Scholar] [CrossRef] [Green Version]
- Mu, M.; An, P.; Wu, Q.; Shen, X.; Shao, D.; Wang, H.; Zhang, Y.; Zhang, S.; Yao, H.; Min, J.; et al. The dietary flavonoid myricetin regulates iron homeostasis by suppressing hepcidin expression. J. Nutr. Biochem. 2016, 30, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Minear, S.; O’Donnell, A.F.; Ballew, A.; Giaever, G.; Nislow, C.; Stearns, T.; Cyert, M.S. Curcumin inhibits growth of Saccharomyces cerevisiae through iron chelation. Eukaryot. Cell 2011, 10, 1574–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, D.; Huebbe, P.; Frank, J.; Rimbach, G.; Pallauf, K. Curcumin may impair iron status when fed to mice for six months. Redox Biol. 2014, 2, 563–569. [Google Scholar] [CrossRef] [Green Version]
- Petry, N. Polyphenols and low iron bioavailability. In Polyphenols in Human Health and Disease; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Academic Press: Oxford, UK, 2014; pp. 311–322. [Google Scholar]
- Kim, E.Y.; Ham, S.; Bradke, D.; Ma, Q.; Han, O. Ascorbic acid offsets the inhibitory effect of bioactive dietary polyphenolic compounds on transepithelial iron transport in Caco-2 intestinal cells. J. Nutr. 2011, 141, 828–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesjak, M.; Balesaria, S.; Skinner, V.; Debnam, E.S.; Srai, S.K.S. Quercetin inhibits intestinal non-haem iron absorption by regulating iron metabolism genes in the tissues. Eur. J. Nutr. 2019, 58, 743–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, H.; Liu, D. Dietary antiaging phytochemicals and mechanisms associated with prolonged survival. J. Nutr. Biochem. 2014, 25, 581–591. [Google Scholar] [CrossRef] [Green Version]
- Kortman, G.A.; Raffatellu, M.; Swinkels, D.W.; Tjalsma, H. Nutritional iron turned inside out: Intestinal stress from a gut Microbial perspective. FEMS Microbiol. Rev. 2014, 38, 1202–1234. [Google Scholar] [CrossRef] [Green Version]
- Jaeggi, T.; Kortman, G.A.; Moretti, D.; Chassard, C.; Holding, P.; Dostal, A.; Boekhorst, J.; Timmerman, H.M.; Swinkels, D.W.; Tjalsma, H.; et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 2015, 64, 731–742. [Google Scholar] [CrossRef]
- Carafa, V.; Rotili, D.; Forgione, M.; Cuomo, F.; Serretiello, E.; Hailu, G.S.; Jarho, E.; Lahtela-Kakkonen, M.; Mai, A.; Altucci, L. Sirtuin functions and modulation: From chemistry to the clinic. Clin. Epigenet. 2016, 8, 61. [Google Scholar] [CrossRef]
- Das, S.K.; DesAulniers, J.; Dyck, J.R.; Kassiri, Z.; Oudit, G.Y. Resveratrol mediates therapeutic hepatic effects in acquired and genetic murine models of iron-overload. Liver Int. 2016, 36, 246–257. [Google Scholar] [CrossRef]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arruda, S.F.; Ramos, L.V.; Barbosa, J.L.d.A.; Hankins, N.A.C.; Rodrigues, P.A.M.; da Cunha, M.d.S.B. The Action of JAK/STAT3 and BMP/HJV/SMAD Signaling Pathways on Hepcidin Suppression by Tucum-do-Cerrado in a Normal and Iron-Enriched Diets. Nutrients 2020, 12, 1515. https://doi.org/10.3390/nu12051515
Arruda SF, Ramos LV, Barbosa JLdA, Hankins NAC, Rodrigues PAM, da Cunha MdSB. The Action of JAK/STAT3 and BMP/HJV/SMAD Signaling Pathways on Hepcidin Suppression by Tucum-do-Cerrado in a Normal and Iron-Enriched Diets. Nutrients. 2020; 12(5):1515. https://doi.org/10.3390/nu12051515
Chicago/Turabian StyleArruda, Sandra Fernandes, Larissa Valadares Ramos, Júlia Lima de Alencar Barbosa, Natália Aboudib Campos Hankins, Pedro Augusto Matos Rodrigues, and Marcela de Sá Barreto da Cunha. 2020. "The Action of JAK/STAT3 and BMP/HJV/SMAD Signaling Pathways on Hepcidin Suppression by Tucum-do-Cerrado in a Normal and Iron-Enriched Diets" Nutrients 12, no. 5: 1515. https://doi.org/10.3390/nu12051515
APA StyleArruda, S. F., Ramos, L. V., Barbosa, J. L. d. A., Hankins, N. A. C., Rodrigues, P. A. M., & da Cunha, M. d. S. B. (2020). The Action of JAK/STAT3 and BMP/HJV/SMAD Signaling Pathways on Hepcidin Suppression by Tucum-do-Cerrado in a Normal and Iron-Enriched Diets. Nutrients, 12(5), 1515. https://doi.org/10.3390/nu12051515