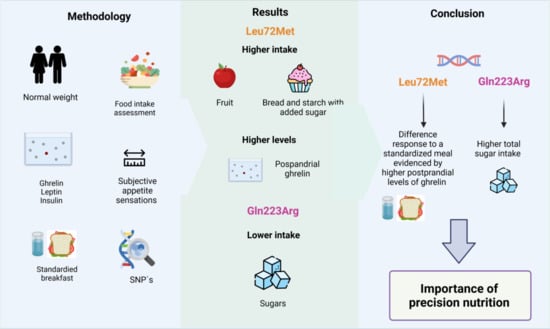
Role of Leu72Met of GHRL and Gln223Arg of LEPR Variants on Food Intake, Subjective Appetite, and Hunger-Satiety Hormones
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Procedure
2.3. Anthropometric Measurements
2.4. Biochemical Determinations
2.5. Quantification of Appetite Hormones
2.6. DNA Extraction and Genotyping
2.7. Dietary Intake
2.8. Breakfast Design
2.9. Appetite Assessment
2.10. Statistical Analysis
3. Results
3.1. Population Description
3.2. Genotypic and Allele Frequencies
3.3. Dietary Intake According to Polymorphisms in GHRL and LEPR Genes
3.4. Subjective Appetite by Polymorphism Leu72Met of GHRL and Gln223Arg of LEPR Genes
3.5. Appetite Hormones
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amin, T.; Mercer, J.G. Hunger and Satiety Mechanisms and Their Potential Exploitation in the Regulation of Food Intake. Curr. Obes. Rep. 2016, 5, 106–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAO. Nutrición Humana en el Mundo en Desarrollo; FAO: Rome, Italy, 2002; ISBN 978-92-5-303818-3. [Google Scholar]
- Flint, A.; Raben, A.; Blundell, J.E.; Astrup, A. Reproducibility, Power and Validity of Visual Analogue Scales in Assessment of Appetite Sensations in Single Test Meal Studies. Int. J. Obes. Relat. Metab. Disord. 2000, 24, 38–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Graaf, C.; Blom, W.A.; Smeets, P.A.; Stafleu, A.; Hendriks, H.F. Biomarkers of Satiation and Satiety. Am. J. Clin. Nutr. 2004, 79, 946–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, J. Hunger, Ghrelin and the Gut. Brain Res. 2018, 1693, 154–158. [Google Scholar] [CrossRef]
- Zhang, Y.; Chua, S. Leptin Function and Regulation. Compr. Physiol. 2017, 8, 351–369. [Google Scholar] [CrossRef]
- Yu, J.H.; Kim, M.-S. Molecular Mechanisms of Appetite Regulation. Diabetes Metab. J. 2012, 36, 391–398. [Google Scholar] [CrossRef]
- Scherer, T.; Sakamoto, K.; Buettner, C. Brain Insulin Signalling in Metabolic Homeostasis and Disease. Nat. Rev. Endocrinol. 2021, 17, 468–483. [Google Scholar] [CrossRef]
- Figlewicz, D.P. Expression of Receptors for Insulin and Leptin in the Ventral Tegmental Area/Substantia Nigra (VTA/SN) of the Rat: Historical Perspective. Brain Res. 2016, 1645, 68–70. [Google Scholar] [CrossRef]
- Isganaitis, E.; Lustig, R.H. Fast Food, Central Nervous System Insulin Resistance, and Obesity. Arter. Thromb. Vasc. Biol. 2005, 25, 2451–2462. [Google Scholar] [CrossRef] [Green Version]
- MacLean, P.S.; Blundell, J.E.; Mennella, J.A.; Batterham, R.L. Biological Control of Appetite: A Daunting Complexity. Obesity 2017, 25, S8–S16. [Google Scholar] [CrossRef] [Green Version]
- Brunner, E.J.; Maruyama, K.; Shipley, M.; Cable, N.; Iso, H.; Hiyoshi, A.; Stallone, D.; Kumari, M.; Tabak, A.; Singh-Manoux, A.; et al. Appetite Disinhibition Rather than Hunger Explains Genetic Effects on Adult BMI Trajectory. Int. J. Obes. 2021, 45, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Crovesy, L.; Rosado, E.L. Interaction between Genes Involved in Energy Intake Regulation and Diet in Obesity. Nutrition 2019, 67–68, 110547. [Google Scholar] [CrossRef] [PubMed]
- Miraglia del Giudice, E.; Santoro, N.; Cirillo, G.; Raimondo, P.; Grandone, A.; D’Aniello, A.; Di Nardo, M.; Perrone, L. Molecular Screening of the Ghrelin Gene in Italian Obese Children: The Leu72Met Variant Is Associated with an Earlier Onset of Obesity. Int. J. Obes. 2004, 28, 447–450. [Google Scholar] [CrossRef] [Green Version]
- Monteleone, P.; Tortorella, A.; Castaldo, E.; Di Filippo, C.; Maj, M. The Leu72Met Polymorphism of the Ghrelin Gene Is Significantly Associated with Binge Eating Disorder. Psychiatr. Genet. 2007, 17, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Suchankova, P.; Yan, J.; Schwandt, M.L.; Stangl, B.L.; Jerlhag, E.; Engel, J.A.; Hodgkinson, C.A.; Ramchandani, V.A.; Leggio, L. The Leu72Met Polymorphism of the Prepro-Ghrelin Gene Is Associated With Alcohol Consumption and Subjective Responses to Alcohol: Preliminary Findings. Alcohol Alcohol. 2017, 52, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Robitaille, J.; Pérusse, L.; Bouchard, C.; Vohl, M.-C. Genes, Fat Intake, and Cardiovascular Disease Risk Factors in the Quebec Family Study. Obesity 2007, 15, 2336–2347. [Google Scholar] [CrossRef]
- Saliba, L.F.; Reis, R.S.; Brownson, R.C.; Hino, A.A.; Tureck, L.V.; Valko, C.; de Souza, R.L.R.; Furtado-Alle, L. Obesity-Related Gene ADRB2, ADRB3 and GHRL Polymorphisms and the Response to a Weight Loss Diet Intervention in Adult Women. Genet. Mol. Biol. 2014, 37, 15–22. [Google Scholar] [CrossRef]
- Takezawa, J.; Yamada, K.; Miyachi, M.; Morita, A.; Aiba, N.; Sasaki, S.; Watanabe, S. Saku Control Obesity Program (SCOP) Study Group Preproghrelin Gene Polymorphisms in Obese Japanese Women. Minor Homozygotes Are Light Eaters, Do Not Prefer Protein or Fat, and Apparently Have a Poor Appetite. Appetite 2013, 63, 105–111. [Google Scholar] [CrossRef]
- Kindler, J.; Bailer, U.; de Zwaan, M.; Fuchs, K.; Leisch, F.; Grün, B.; Strnad, A.; Stojanovic, M.; Windisch, J.; Lennkh-Wolfsberg, C.; et al. No Association of the Neuropeptide Y (Leu7Pro) and Ghrelin Gene (Arg51Gln, Leu72Met, Gln90Leu) Single Nucleotide Polymorphisms with Eating Disorders. Nord. J. Psychiatry 2011, 65, 203–207. [Google Scholar] [CrossRef]
- Daghestani, M.; Purohit, R.; Daghestani, M.; Daghistani, M.; Warsy, A. Molecular Dynamic (MD) Studies on Gln233Arg (Rs1137101) Polymorphism of Leptin Receptor Gene and Associated Variations in the Anthropometric and Metabolic Profiles of Saudi Women. PLoS ONE 2019, 14, e0211381. [Google Scholar] [CrossRef]
- Nesrine, Z.; Haithem, H.; Imen, B.; Fadoua, N.; Asma, O.; Fadhel, N.M.; Ali, B. Leptin and Leptin Receptor Polymorphisms, Plasma Leptin Levels and Obesity in Tunisian Volunteers. Int. J. Exp. Pathol. 2018, 99, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Rojano-Rodriguez, M.E.; Beristain-Hernandez, J.L.; Zavaleta-Villa, B.; Maravilla, P.; Romero-Valdovinos, M.; Olivo-Diaz, A. Leptin Receptor Gene Polymorphisms and Morbid Obesity in Mexican Patients. Hereditas 2016, 153, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boumaiza, I.; Omezzine, A.; Rejeb, J.; Rebhi, L.; Ouedrani, A.; Ben Rejeb, N.; Nabli, N.; Ben Abdelaziz, A.; Bouslama, A. Relationship between Leptin G2548A and Leptin Receptor Q223R Gene Polymorphisms and Obesity and Metabolic Syndrome Risk in Tunisian Volunteers. Genet. Test. Mol. Biomark. 2012, 16, 726–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, A.D.; McGregor, J.C.; Perencevich, E.N.; Furuno, J.P.; Zhu, J.; Peterson, D.E.; Finkelstein, J. The Use and Interpretation of Quasi-Experimental Studies in Medical Informatics. J. Am. Med. Inform. Assoc. 2006, 13, 16–23. [Google Scholar] [CrossRef]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, without Use of the Preparative Ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef]
- Blundell, J.; de Graaf, C.; Hulshof, T.; Jebb, S.; Livingstone, B.; Lluch, A.; Mela, D.; Salah, S.; Schuring, E.; van der Knaap, H.; et al. Appetite Control: Methodological Aspects of the Evaluation of Foods. Obes. Rev. 2010, 11, 251–270. [Google Scholar] [CrossRef] [Green Version]
- Korek, E.; Krauss, H.; Gibas-Dorna, M.; Kupsz, J.; Piątek, M.; Piątek, J. Fasting and Postprandial Levels of Ghrelin, Leptin and Insulin in Lean, Obese and Anorexic Subjects. Prz. Gastroenterol. 2013, 8, 383–389. [Google Scholar] [CrossRef]
- Tschöp, M.; Weyer, C.; Tataranni, P.A.; Devanarayan, V.; Ravussin, E.; Heiman, M.L. Circulating Ghrelin Levels Are Decreased in Human Obesity. Diabetes 2001, 50, 707–709. [Google Scholar] [CrossRef] [Green Version]
- Espinoza-García, A.S.; Hunot-Alexander, C.; Martínez-Moreno, A.G.; Vázquez-Solorzano, R.; Porchas-Quijada, M.; Reyes-Castillo, Z. IgG Antibodies Reacting with Ghrelin and Leptin Are Correlated with Body Composition and Appetitive Traits in Young Subjects. Appetite 2022, 168, 105685. [Google Scholar] [CrossRef]
- Tobin, S.Y.; Cornier, M.-A.; White, M.H.; Hild, A.K.; Simonsen, S.E.; Melanson, E.L.; Halliday, T.M. The Effects of Acute Exercise on Appetite and Energy Intake in Men and Women. Physiol. Behav. 2021, 241, 113562. [Google Scholar] [CrossRef]
- Ali, E.M.M.; Diab, T.; Elsaid, A.; Abd El Daim, H.A.; Elshazli, R.M.; Settin, A. Fat Mass and Obesity-Associated (FTO) and Leptin Receptor (LEPR) Gene Polymorphisms in Egyptian Obese Subjects. Arch. Physiol. Biochem. 2021, 127, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Rs696217 (SNP)-Population Genetics-Homo_sapiens-Ensembl Genome Browser 105. Available online: http://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=3:10289273-10290273;v=rs696217;vdb=variation;vf=90594079 (accessed on 10 April 2022).
- Rivera-León, E.A.; Llamas-Covarrubias, M.A.; Sánchez-Enríquez, S.; Martínez-López, E.; González-Hita, M.; Llamas-Covarrubias, I.M. Leu72Met Polymorphism of GHRL Gene Decreases Susceptibility to Type 2 Diabetes Mellitus in a Mexican Population. BMC Endocr. Disord. 2020, 20, 109. [Google Scholar] [CrossRef] [PubMed]
- Rs1137101 (SNP)-Population Genetics-Homo_sapiens-Ensembl Genome Browser 105. Available online: http://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=1:65592330-65593330;v=rs1137101;vdb=variation;vf=841289 (accessed on 10 April 2022).
- Adamska-Patruno, E.; Ostrowska, L.; Goscik, J.; Pietraszewska, B.; Kretowski, A.; Gorska, M. The Relationship between the Leptin/Ghrelin Ratio and Meals with Various Macronutrient Contents in Men with Different Nutritional Status: A Randomized Crossover Study. Nutr. J. 2018, 17, 118. [Google Scholar] [CrossRef] [Green Version]
- Rolls, B.J. Carbohydrates, Fats, and Satiety. Am. J. Clin. Nutr. 1995, 61, 960S–967S. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, P.; Bencivenga, R.; Longobardi, N.; Serritella, C.; Maj, M. Differential Responses of Circulating Ghrelin to High-Fat or High-Carbohydrate Meal in Healthy Women. J. Clin. Endocrinol. Metab. 2003, 88, 5510–5514. [Google Scholar] [CrossRef]
- Vitolo, E.; Santini, E.; Seghieri, M.; Giannini, L.; Coppedè, F.; Rossi, C.; Dardano, A.; Solini, A. Heterozygosity for the Rs696217 SNP in the Preproghrelin Gene Predicts Weight Loss After Bariatric Surgery in Severely Obese Individuals. Obes. Surg. 2017, 27, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Ukkola, O.; Ravussin, E.; Jacobson, P.; Pérusse, L.; Rankinen, T.; Tschöp, M.; Heiman, M.L.; Leon, A.S.; Rao, D.C.; Skinner, J.S.; et al. Role of Ghrelin Polymorphisms in Obesity Based on Three Different Studies. Obes. Res. 2002, 10, 782–791. [Google Scholar] [CrossRef] [Green Version]
- Hedayatizadeh-Omran, A.; Rafiei, A.; Khajavi, R.; Alizadeh-Navaei, R.; Mokhberi, V.; Moradzadeh, K. Association between Ghrelin Gene (Leu72Met) Polymorphism and Ghrelin Serum Level with Coronary Artery Diseases. DNA Cell Biol. 2014, 33, 95–101. [Google Scholar] [CrossRef]
- Domínguez-Reyes, T.; Astudillo-López, C.C.; Salgado-Goytia, L.; Muñoz-Valle, J.F.; Salgado-Bernabé, A.B.; Guzmán-Guzmán, I.P.; Castro-Alarcón, N.; Moreno-Godínez, M.E.; Parra-Rojas, I. Interaction of Dietary Fat Intake with APOA2, APOA5 and LEPR Polymorphisms and Its Relationship with Obesity and Dyslipidemia in Young Subjects. Lipids Health Dis. 2015, 14, 106. [Google Scholar] [CrossRef] [Green Version]
- Mizuta, E.; Kokubo, Y.; Yamanaka, I.; Miyamoto, Y.; Okayama, A.; Yoshimasa, Y.; Tomoike, H.; Morisaki, H.; Morisaki, T. Leptin Gene and Leptin Receptor Gene Polymorphisms Are Associated with Sweet Preference and Obesity. Hypertens. Res. 2008, 31, 1069–1077. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, R.; Noguchi, K.; Shigemura, N.; Jyotaki, M.; Takahashi, I.; Margolskee, R.F.; Ninomiya, Y. Leptin Suppresses Mouse Taste Cell Responses to Sweet Compounds. Diabetes 2015, 64, 3751–3762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, K.; Sugimoto, K.; Nakashima, K.; Miura, H.; Ninomiya, Y. Leptin as a Modulator of Sweet Taste Sensitivities in Mice. Proc. Natl. Acad. Sci. USA 2000, 97, 11044–11049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, B.; Breza, J.M.; Nikonov, A.A.; Paedae, A.B.; Contreras, R.J. Leptin Increases Temperature-Dependent Chorda Tympani Nerve Responses to Sucrose in Mice. Physiol. Behav. 2012, 107, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Sanematsu, K.; Ohta, R.; Shirosaki, S.; Koyano, K.; Nonaka, K.; Shigemura, N.; Ninomiya, Y. Diurnal Variation of Human Sweet Taste Recognition Thresholds Is Correlated with Plasma Leptin Levels. Diabetes 2008, 57, 2661–2665. [Google Scholar] [CrossRef] [Green Version]
- Park, K.S.; Shin, H.D.; Park, B.L.; Cheong, H.S.; Cho, Y.M.; Lee, H.K.; Lee, J.-Y.; Lee, J.-K.; Oh, B.; Kimm, K. Polymorphisms in the Leptin Receptor (LEPR)--Putative Association with Obesity and T2DM. J. Hum. Genet. 2006, 51, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Valente, C.; Alvarez, L.; Marques, P.I.; Gusmão, L.; Amorim, A.; Seixas, S.; João Prata, M. Genes from the TAS1R and TAS2R Families of Taste Receptors: Looking for Signatures of Their Adaptive Role in Human Evolution. Genome Biol. Evol. 2018, 10, 1139–1152. [Google Scholar] [CrossRef] [Green Version]
- Aukan, M.I.; Nymo, S.; Haagensli Ollestad, K.; Akersveen Boyesen, G.; DeBenedictis, J.N.; Rehfeld, J.F.; Coutinho, S.; Martins, C. Differences in Gastrointestinal Hormones and Appetite Ratings among Obesity Classes. Appetite 2022, 171, 105940. [Google Scholar] [CrossRef]
- Joannic, J.L.; Oppert, J.M.; Lahlou, N.; Basdevant, A.; Auboiron, S.; Raison, J.; Bornet, F.; Guy-Grand, B. Plasma Leptin and Hunger Ratings in Healthy Humans. Appetite 1998, 30, 129–138. [Google Scholar] [CrossRef]
- Cruwys, T.; Bevelander, K.E.; Hermans, R.C.J. Social Modeling of Eating: A Review of When and Why Social Influence Affects Food Intake and Choice. Appetite 2015, 86, 3–18. [Google Scholar] [CrossRef]





| Variables | All n = 132 | Women n = 102 | Men n = 30 | p-Value |
|---|---|---|---|---|
| Anthropometric parameters | ||||
| BMI (kg/m2) | 22.0 ± 2.0 | 21.7 ± 1.9 | 22.9 ± 2.1 | 0.005 |
| WC (cm) | 71.6 ± 6.4 | 69.6 ± 4.9 | 78.3 ± 6.2 | 0.005 |
| Fat mass (kg) | 17 ± 5.8 | 17.4 ± 4.5 | 16.2 ± 8.8 | <0.001 |
| BFP (%) | 28.4 ± 7.1 | 30.4 ± 6.0 | 21.6 ± 6.4 | <0.001 |
| FFM (kg) | 42.8 ± 8.3 | 39.5 ± 5.3 | 54.1 ± 6.7 | <0.001 |
| Lean mass (kg) | 42.6 ± 28.8 | 40.1 ± 32.2 | 51.1 ± 6.3 | <0.001 |
| SMM (kg) | 23.4 ± 5.1 | 21.3 ± 3.2 | 30.4 ± 4.0 | <0.001 |
| Mineral mass (kg) | 3.0 ± 0.6 | 2.8 ± 0.4 | 3.7 ± 0.5 | <0.001 |
| TBW (kg) | 31.3 ± 6.1 | 28.8 ± 3.8 | 39.7 ± 4.9 | <0.001 |
| Systolic blood pressure (mmHg) | 109.4 ± 10.5 | 105.8 ± 8.3 | 121.5 ± 7.8 | <0.001 |
| Diastolic blood pressure (mmHg) | 66.8 ± 7.2 | 66.0 ± 7.3 | 69.3 ± 6.5 | 0.028 |
| Biochemical parameters | ||||
| TC (mg/dL) | 146.7 ± 27.9 | 146.8 ± 28.2 | 146.3 ± 26.9 | 0.929 |
| HDL-C (mg/dL) | 49.7 ± 11.9 | 50.4 ± 11.8 | 46.8 ± 11.9 | 0.147 |
| LDL-C (mg/dL) | 80.3 ± 22.4 | 79.6 ± 23.0 | 82.4 ± 20.5 | 0.545 |
| VLDL-C (mg/dL) | 15.9 ± 5.9 | 15.8 ± 5.7 | 85.3 ± 34.2 | 0.294 |
| Triglycerides (mg/dL) | 80.0 ± 29.2 | 79.0 ± 28.2 | 46.8 ± 11.9 | 0.270 |
| Glucose (mg/dL) | 90.9 ± 11.9 | 90.4 ± 10.2 | 92.6 ± 10.7 | 0.366 |
| Insulin (µUI/mL) | 7.2 ± 4.2 | 7.4 ± 4.0 | 6.7 ± 4.8 | 0.395 |
| HOMA-IR | 1.6 ± 1.0 | 1.7 ± 1.0 | 1.5 ± 1.1 | 0.960 |
| Ghrelin (pg/mL) | 511.1 ± 346.5 | 466.5 ± 329.8 | 658.6 ± 364.8 | 0.001 |
| Leptin (ng/mL) | 11.1 ± 6.4 | 11.5 ± 6.6 | 10.0 ± 5.7 | 0.275 |
| Others | ||||
| Kilocalories from dinner one day before intervention (kcal) | 457.6 ± 301.1 | 448.2 ± 286.2 | 483.1 ± 344.3 | 0.674 |
| Carbohydrates from dinner one day before intervention (%) | 53.5 ± 19.1 | 56.2 ± 19.1 | 46.2 ± 17.5 | 0.035 |
| Protein from dinner one day before intervention (%) | 18.2 ± 9.7 | 17.7 ± 9.4 | 19.5 ± 10.3 | 0.476 |
| Fat, total from dinner one day before intervention (%) | 30.8 ± 14.5 | 28.8 ± 14.3 | 36.5 ± 13.7 | 0.031 |
| Available carbohydrate from dinner one day before intervention (g) | 1.1 ± 3.5 | 0.6 ± 2.8 | 2.5 ± 4.9 | 0.096 |
| Alcohol from dinner one day before intervention (g) | 0.02 ± 0.2 | 0.03 ± 0.2 | 0.0 ± 0.0 | 0.449 |
| Dinner time one day before intervention (h) | 21:23 ± 1:13 | 21:15 ± 1:13 | 21:3 ± 1:12 | 0.137 |
| Leu72Met of GHRL | n (%) | |
| Genotype | Leu/Leu | 120 (91) |
| Leu/Met | 12 (9) | |
| Met/Met | 0 (0) | |
| HWE p = 0.5843 | ||
| Allele | Leu | 252 (95) |
| Met | 12 (5) | |
| Dominant model | Leu/Leu | 120 (91) |
| Leu/Met + Met/Met | 12 (9) | |
| Gln223Arg of LEPR | n (%) | |
| Genotype | Gln/Gln | 37 (28) |
| Gln/Arg | 68 (51) | |
| Arg/Arg | 27 (21) | |
| HWE p = 0.8070 | ||
| Allele | Gln | 142 (54) |
| Arg | 122 (46) | |
| Dominant model | Gln/Gln | 37 (28) |
| Gln/Arg + Arg/Arg | 95 (72) | |
| Variable | Leu/Leu n = 120 | Leu/Met + Met/Met n = 12 | p-Value | Gln/Gln n = 37 | Gln/Arg + Arg/Arg n = 95 | p-Value |
|---|---|---|---|---|---|---|
| Sugar total (g/d) | 83.5 (74.6–92.4) | 103.4 (75.4–131.4) | 0.183 | 101.0 (85.2–116.9) | 79.0 (68.9–89.0) | 0.021 |
| Fruit servings/d | 1.9 (1.6–2.2) | 2.9 (2.0–3.8) | 0.045 | 2.3 (1.8–2.9) | 1.9 (1.6–2.2) | 0.166 |
| Bread/starch with added sugar servings/d | 2.1 (1.7–2.5) | 3.5 (2.2–4.8) | 0.043 | 2.7 (2.0–3.4) | 1.9 (1.5–2.4) | 0.086 |
| Hormones | All n = 132 | Leu/Leu n = 120 | Leu/Met + Met/Met n = 12 | p-Value | Gln/Gln n = 37 | Gln/Arg + Arg/Arg n = 95 | p-Value |
|---|---|---|---|---|---|---|---|
| Basal ghrelin (pg/mL) | 511.1 ± 346.5 378.7 (317.9–653.5) | 502.0 ± 340.4 377.3 (318.0–600.8) | 600.2 ± 406.8 635.5 (287.5–726.8) | 0.313 | 509.6 ± 420.4 366.0 (244.0–652.0) | 514.4 ± 320.2 378.4 (330.0–692.5) | 0.264 |
| Final ghrelin (pg/mL) | 420.9 ± 280.0 354.0 (272.8–475.0) | 407.3 ± 264.6 350.0 (271.9–462.6) | 556.1 ± 392.0 440.7 (347.2–701.6) | 0.079 | 423.0 ± 394.8 321.7 (234.5–409.1) | 419.0 ± 223.9 357.2 (276.3–494.7) | 0.103 |
| Basal leptin (ng/mL) | 11.1 ± 6.3 10.8 (7.0–15.8) | 11.0 ± 6.3 10.8 (7.0–15.0) | 12.4 ± 7.8 11.3 (5.7–20.0) | 0.534 | 12.4 ± 5.5 13.0 (8.0–17.0) | 10.5 ± 6.6 10.0 (5.7–14.2) | 0.035 |
| Final leptin (ng/mL) | 9.1± 6.4 7.9 (4.2–14.0) | 9.1± 6.3 7.9 (4.2–14.0) | 9.5 ± 7.9 7.0 (2.8–12.9) | 0.889 | 10.4 ± 6.5 8.0 (5.0–14.0) | 8.5 ± 6.3 6.3 (3.1–12.9) | 0.085 |
| Basal insulin (µUI/mL) | 7.2 ± 4.2 6.1 (4.7–8.6) | 7.2 ± 4.2 5.9 (4.5–8.4) | 8.4 ± 4.5 6.9 (4.9–12.2) | 0.379 | 7.5 ± 3.7 6.8 (4.9–9.1) | 7.2 ± 4.5 6.0 (4.5–8.2) | 0.403 |
| Final insulin (µUI/mL) | 12.6 ± 8.6 10.1 (6.7–15.9) | 12.6 ± 8.8 10.0 (6.6–15.6) | 12.6 ± 7.7 10.4 (7.2–20.7) | 0.772 | 14.2 ± 12.0 9.4 (6.8–19.5) | 11.9 ± 7.2 10.2 (6.5–15.1) | 0.908 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Murguia, T.; Torres-Castillo, N.; Magaña-de la Vega, L.; Rodríguez-Reyes, S.C.; Campos-Pérez, W.; Martínez-López, E. Role of Leu72Met of GHRL and Gln223Arg of LEPR Variants on Food Intake, Subjective Appetite, and Hunger-Satiety Hormones. Nutrients 2022, 14, 2100. https://doi.org/10.3390/nu14102100
Sanchez-Murguia T, Torres-Castillo N, Magaña-de la Vega L, Rodríguez-Reyes SC, Campos-Pérez W, Martínez-López E. Role of Leu72Met of GHRL and Gln223Arg of LEPR Variants on Food Intake, Subjective Appetite, and Hunger-Satiety Hormones. Nutrients. 2022; 14(10):2100. https://doi.org/10.3390/nu14102100
Chicago/Turabian StyleSanchez-Murguia, Tania, Nathaly Torres-Castillo, Lisset Magaña-de la Vega, Saraí Citlalic Rodríguez-Reyes, Wendy Campos-Pérez, and Erika Martínez-López. 2022. "Role of Leu72Met of GHRL and Gln223Arg of LEPR Variants on Food Intake, Subjective Appetite, and Hunger-Satiety Hormones" Nutrients 14, no. 10: 2100. https://doi.org/10.3390/nu14102100
APA StyleSanchez-Murguia, T., Torres-Castillo, N., Magaña-de la Vega, L., Rodríguez-Reyes, S. C., Campos-Pérez, W., & Martínez-López, E. (2022). Role of Leu72Met of GHRL and Gln223Arg of LEPR Variants on Food Intake, Subjective Appetite, and Hunger-Satiety Hormones. Nutrients, 14(10), 2100. https://doi.org/10.3390/nu14102100