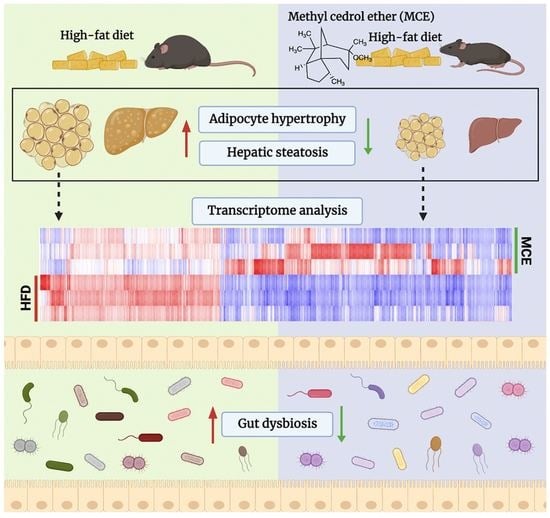
Dietary Supplementation of Methyl Cedryl Ether Ameliorates Adiposity in High-Fat Diet-Fed Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animals and Designs
2.3. Fasting Blood Glucose (FBG) and Biochemical Parameters Analysis
2.4. Histological Examination
2.5. Analysis of Hepatic Triglyceride (TG) and TC Content
2.6. Real-Time Quantitative PCR (RT-qPCR)
2.7. Gut Microbiota Analysis
2.8. Epididymal WAT Transcriptome Analysis
2.9. Association Analysis of Gut Microbiota and DEGs
2.10. Statistical Analysis
3. Results
3.1. MCE Has a Strong Preventive Potential against HFD-Induced Body Weight Gain in Mice

3.2. MCE Lowers FBG and Improves Serum Lipid Profile in HFD-Fed Mice
3.3. MCE Attenuates the Adipocyte Hypertrophy and Hepatic Steatosis in HFD-Fed Mice
3.4. MCE Alters the Expression of Genes Involved in Adipogenesis and Thermogenesis in Epididymal WAT
3.5. MCE-Supplemented Mice Show Apparent Changes in Gut Microbial Structure
3.6. Predicted Metabolic Profiles of the Gut Microbiota after MCE Supplementation
3.7. MCE Supplementation Modifies the Epididymal WAT Transcriptome in HFD-Fed Mice
3.8. Interactions between Gene Expression Profile in WAT and Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Gonzalez-Muniesa, P.; Martinez-Gonzalez, M.A.; Hu, F.B.; Despres, J.P.; Matsuzawa, Y.; Loos, R.J.F.; Moreno, L.A.; Bray, G.A.; Martinez, J.A. Obesity. Nat. Rev. Dis. Prim. 2017, 3, 17034. [Google Scholar] [CrossRef] [PubMed]
- Piche, M.E.; Tchernof, A.; Despres, J.P. Obesity Phenotypes, Diabetes, and Cardiovascular Diseases. Circ. Res. 2020, 126, 1477–1500. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.Z.; Garske, K.M.; Alvarez, M.; Bhagat, Y.V.; Boocock, J.; Nikkola, E.; Miao, Z.; Raulerson, C.K.; Cantor, R.M.; Civelek, M.; et al. Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS. Nat. Commun. 2018, 9, 1512. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R.; Olefsky, J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 2017, 127, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Lin, L. Small molecules for fat combustion: Targeting obesity. Acta Pharm. Sin. B 2019, 9, 220–236. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, N.C.; Groen, A.K.; Romijn, J.A.; Nieuwdorp, M. Gut Microbiota in Obesity and Undernutrition. Adv. Nutr. 2016, 7, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.; Wang, Y.; Geng, R.; Fang, J.; Liu, Q.; Kang, S.-G.; Zeng, W.C.; Huang, K.; Tong, T. Understanding the mechanism underlying the anti-diabetic effect of dietary component: A focus on gut microbiota. Crit. Rev. Food Sci. Nutr. 2022, 1–21. [Google Scholar] [CrossRef]
- Ruan, J.-W.; Statt, S.; Huang, C.-T.; Tsai, Y.-T.; Kuo, C.-C.; Chan, H.-L.; Liao, Y.-C.; Tan, T.-H.; Kao, C.-Y. Dual-specificity phosphatase 6 deficiency regulates gut microbiome and transcriptome response against diet-induced obesity in mice. Nat. Microbiol. 2016, 2, 16220. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, Y.; Fang, J.; Geng, R.; Li, M.; Zhao, Y.; Kang, S.-G.; Huang, K.; Tong, T. Oleuropein Ameliorates Advanced Stage of Type 2 Diabetes in db/db Mice by Regulating Gut Microbiota. Nutrients 2021, 13, 2131. [Google Scholar] [CrossRef]
- Torres-Fuentes, C.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. The microbiota-gut-brain axis in obesity. Lancet Gastroenterol. Hepatol. 2017, 2, 747–756. [Google Scholar] [CrossRef]
- Li, M.; Zhao, Y.; Wang, Y.; Geng, R.; Fang, J.; Kang, S.G.; Huang, K.; Tong, T. Eugenol, A Major Component of Clove Oil, Attenuates Adiposity, and Modulates Gut Microbiota in High-Fat Diet-Fed Mice. Mol. Nutr. Food Res. 2022, 66, e2200387. [Google Scholar] [CrossRef]
- Geng, R.; Wang, Y.; Fang, J.; Zhao, Y.; Li, M.; Kang, S.-G.; Huang, K.; Tong, T. Ectopic odorant receptors responding to flavor compounds in skin health and disease: Current insights and future perspectives. Crit. Rev. Food Sci. Nutr. 2022, 1–17. [Google Scholar] [CrossRef]
- Kaur, J.; Gulati, M.; Singh, S.K.; Kuppusamy, G.; Kapoor, B.; Mishra, V.; Gupta, S.; Arshad, M.F.; Porwal, O.; Jha, N.K.; et al. Discovering multifaceted role of vanillic acid beyond flavours: Nutraceutical and therapeutic potential. Trends Food Sci. Technol. 2022, 122, 187–200. [Google Scholar] [CrossRef]
- Chen, X.; Shen, J.; Zhao, J.-M.; Guan, J.; Li, W.; Xie, Q.-M.; Zhao, Y.-Q. Cedrol attenuates collagen-induced arthritis in mice and modulates the inflammatory response in LPS-mediated fibroblast-like synoviocytes. Food Funct. 2020, 11, 4752–4764. [Google Scholar] [CrossRef]
- Batur, O.O.; Kiran, I.; Demirci, F.; Başer, K.H.C. Fungal biotransformation of cedramber. Biocatal. Biotransfor. 2022, 40, 248–251. [Google Scholar] [CrossRef]
- Sağır, T. Halofilik Mikroorganizmalar Ile Biyoaktif Metabolit Üretimi. Ph.D. Thesis, Anadolu University (Turkey), Ann Arbor, MI, USA, 2014. [Google Scholar]
- Gross, A.D. Expression of the Periplaneta americana’s α-Adrenergic-like Octopamine Receptor in the Yeast Saccharomyces cerevisiae: A High-Throughput Screening System in Search of Biorational Insecticides. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2010. [Google Scholar]
- Tong, T.; Yu, R.; Park, T. α-Cedrene protects rodents from high-fat diet-induced adiposity via adenylyl cyclase 3. Int. J. Obes. 2019, 43, 202–216. [Google Scholar] [CrossRef]
- Tong, T.; Ryu, S.E.; Min, Y.; de March, C.A.; Bushdid, C.; Golebiowski, J.; Moon, C.; Park, T. Olfactory receptor 10J5 responding to alpha-cedrene regulates hepatic steatosis via the cAMP-PKA pathway. Sci. Rep. 2017, 7, 9471. [Google Scholar] [CrossRef]
- Kim, Y.J.; Park, T. Genes are differentially expressed in the epididymal fat of rats rendered obese by a high-fat diet. Nutr. Res. 2008, 28, 414–422. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; He, X.; Sheng, Y.; Yang, C.; Li, H.; Xu, J.; Xu, W.; Huang, K. Caulis Spatholobi Ameliorates Obesity through Activating Brown Adipose Tissue and Modulating the Composition of Gut Microbiota. Int. J. Mol. Sci. 2019, 20, 5150. [Google Scholar] [CrossRef]
- Tchernof, A.; Després, J.-P. Pathophysiology of Human Visceral Obesity: An Update. Physiol. Rev. 2013, 93, 359–404. [Google Scholar] [CrossRef] [Green Version]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. 2008, 22, 659–661. [Google Scholar] [CrossRef]
- Cedrol methyl ether. Food Cosmet. Toxicol. 1979, 17, 747. [CrossRef]
- Boby, N.; Abbas, M.A.; Lee, E.-B.; Im, Z.-E.; Lee, S.-J.; Park, S.-C. Microbiota modulation and anti-obesity effects of fermented Pyrus ussuriensis Maxim extract against high-fat diet-induced obesity in rats. Biomed. Pharmacother. 2022, 154, 113629. [Google Scholar] [CrossRef]
- Islam, T.; Koboziev, I.; Albracht-Schulte, K.; Mistretta, B.; Scoggin, S.; Yosofvand, M.; Moussa, H.; Zabet-Moghaddam, M.; Ramalingam, L.; Gunaratne, P.H.; et al. Curcumin Reduces Adipose Tissue Inflammation and Alters Gut Microbiota in Diet-Induced Obese Male Mice. Mol. Nutr. Food Res. 2021, 65, e2100274. [Google Scholar] [CrossRef]
- Du, J.; Zhang, P.; Luo, J.; Shen, L.; Zhang, S.; Gu, H.; He, J.; Wang, L.; Zhao, X.; Gan, M.; et al. Dietary betaine prevents obesity through gut microbiota-drived microRNA-378a family. Gut Microbes 2021, 13, 1862612. [Google Scholar] [CrossRef]
- Lee, P.S.; Teng, C.Y.; Kalyanam, N.; Ho, C.T.; Pan, M.H. Garcinol Reduces Obesity in High-Fat-Diet-Fed Mice by Modulating Gut Microbiota Composition. Mol. Nutr. Food Res. 2019, 63, e1800390. [Google Scholar] [CrossRef]
- Chang, C.-J.; Lin, C.-S.; Lu, C.-C.; Martel, J.; Ko, Y.-F.; Ojcius, D.M.; Tseng, S.-F.; Wu, T.-R.; Chen, Y.-Y.M.; Young, J.D.; et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 2015, 6, 7489. [Google Scholar] [CrossRef]
- Liu, J.; Zhen, D.; Hu, C.; Liu, Y.; Shen, X.; Fu, P.; He, Y. Reconfiguration of Gut Microbiota and Reprogramming of Liver Metabolism with Phycobiliproteins Bioactive Peptides to Rehabilitate Obese Rats. Nutrients 2022, 14, 3635. [Google Scholar] [CrossRef]
- Langwig, M.V.; De Anda, V.; Dombrowski, N.; Seitz, K.W.; Rambo, I.M.; Greening, C.; Teske, A.P.; Baker, B.J. Large-scale protein level comparison of Deltaproteobacteria reveals cohesive metabolic groups. ISME J. 2022, 16, 307–320. [Google Scholar] [CrossRef]
- Cai, T.-T.; Ye, X.-L.; Li, R.-R.; Chen, H.; Wang, Y.-Y.; Yong, H.-J.; Pan, M.-L.; Lu, W.; Tang, Y.; Miao, H.; et al. Resveratrol Modulates the Gut Microbiota and Inflammation to Protect Against Diabetic Nephropathy in Mice. Front. Pharmacol. 2020, 11, 1249. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Yan, S.; Chen, J.; Zhu, L.; Guo, T.; Qin, D.; Hu, Z.; Han, S.; Wang, J.; Matias, F.B.; Wen, L.; et al. Oryzanol alleviates high fat and cholesterol diet-induced hypercholesterolemia associated with the modulation of the gut microbiota in hamsters. Food Funct. 2022, 13, 4486–4501. [Google Scholar] [CrossRef]
- Lyu, Q.; Chen, L.; Lin, S.; Cao, H.; Teng, H. A designed self-microemulsion delivery system for dihydromyricetin and its dietary intervention effect on high-fat-diet fed mice. Food Chem. 2022, 390, 132954. [Google Scholar] [CrossRef]
- Chen, H.; Sun, Y.; Zhao, H.; Qi, X.; Cui, H.; Li, Q.; Ma, Y. α-Lactalbumin peptide Asp-Gln-Trp alleviates hepatic insulin resistance and modulates gut microbiota dysbiosis in high-fat diet-induced NAFLD mice. Food Funct. 2022, 13, 9878–9892. [Google Scholar] [CrossRef]
- Jeremic, N.; Chaturvedi, P.; Tyagi, S.C. Browning of White Fat: Novel Insight Into Factors, Mechanisms, and Therapeutics. J. Cell. Physiol. 2017, 232, 61–68. [Google Scholar] [CrossRef]
- Sarjeant, K.; Stephens, J.M. Adipogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008417. [Google Scholar] [CrossRef]
- Tong, T.; Shen, Y.; Lee, H.-W.; Yu, R.; Park, T. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Sci. Rep. 2016, 6, srep34179. [Google Scholar] [CrossRef]
- Lee, H.S.; Lim, S.-M.; Jung, J.I.; Kim, S.M.; Lee, J.K.; Kim, Y.H.; Cha, K.M.; Oh, T.K.; Moon, J.M.; Kim, T.Y.; et al. Gynostemma Pentaphyllum Extract Ameliorates High-Fat Diet-Induced Obesity in C57BL/6N Mice by Upregulating SIRT1. Nutrients 2019, 11, 2475. [Google Scholar] [CrossRef]
- Tong, T.; Park, J.; Moon, C.; Park, T. Regulation of Adipogenesis and Thermogenesis through Mouse Olfactory Receptor 23 Stimulated by α-Cedrene in 3T3-L1 Cells. Nutrients 2018, 10, 1781. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, N.; Tan, H.Y.; Li, S.; Zhang, C.; Zhang, Z.; Feng, Y. Panax notoginseng saponins modulate the gut microbiota to promote thermogenesis and beige adipocyte reconstruction via leptin-mediated AMPKalpha/STAT3 signaling in diet-induced obesity. Theranostics 2020, 10, 11302–11323. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Li, B.; Meng, X.; Wang, J.; Zhang, Y.; Yao, S.; Ma, Q.; Jin, L.; Yang, J.; et al. Berberine activates thermogenesis in white and brown adipose tissue. Nat. Commun. 2014, 5, 5493. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Liu, Q.; Li, Y.; Li, H.; Chen, L.; Yang, X.; Tang, Q.; Pu, S.; Kuang, J.; Li, R.; et al. Feeding-induced hepatokine, Manf, ameliorates diet-induced obesity by promoting adipose browning via p38 MAPK pathway. J. Exp. Med. 2021, 218, e20201203. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Liu, J.; Wang, B.; Zhang, C.; Su, Z.; Zhao, M.; Qin, L.; Zhang, W.; Zheng, R. Withaferin A promotes white adipose browning and prevents obesity through sympathetic nerve-activated Prdm16-FATP1 axis. Diabetes 2022, 71, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shao, M.; Xiang, H.; Zheng, P.; Wu, T.; Ji, G. Integrative transcriptomics and metabolomics explore the mechanism of kaempferol on improving nonalcoholic steatohepatitis. Food Funct. 2020, 11, 10058–10069. [Google Scholar] [CrossRef] [PubMed]
- Benard, O.; Lim, J.; Apontes, P.; Jing, X.; Angeletti, R.H.; Chi, Y. Impact of high-fat diet on the proteome of mouse liver. J. Nutr. Biochem. 2016, 31, 10–19. [Google Scholar] [CrossRef]
- Fabbrini, E.; Sullivan, S.; Klein, S. Obesity and nonalcoholic fatty liver disease: Biochemical, metabolic, and clinical implications. Hepatology 2010, 51, 679–689. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Kountouras, J.; Mantzoros, C.S. Obesity and nonalcoholic fatty liver disease: From pathophysiology to therapeutics. Metabolism 2019, 92, 82–97. [Google Scholar] [CrossRef]






| Ingredients | HFD Group | MCE Group |
|---|---|---|
| Casein | 200 | 200 |
| DL-Methionine | 3 | 3 |
| Corn starch | 111 | 109 |
| Sucrose | 370 | 370 |
| Cellulose | 50 | 50 |
| Corn oil | 30 | 30 |
| Lard | 170 | 170 |
| Mineral mixture a | 42 | 42 |
| Vitamin mixture b | 12 | 12 |
| Choline bitartrate | 2 | 2 |
| Cholesterol | 10 | 10 |
| Methyl cedryl ether | - | 2 |
| tert-Butylhydroquinone c | 0.04 | 0.04 |
| Fat, % kJ | 40 | 40 |
| Chow | HFD | MCE | |
|---|---|---|---|
| FBG (mmol/L) | 8.41 ± 0.22 | 10.01 ± 0.46 ** | 7.68 ± 0.51 ## |
| TC (mmol/L) | 2.83 ± 0.08 | 7.30 ± 0.31 **** | 4.29 ± 0.16 #### |
| HDL-C (mmol/L) | 2.05 ± 0.07 | 3.57 ± 0.11 **** | 2.68 ± 0.14 ### |
| LDL-C (mmol/L) | 0.34 ± 0.01 | 1.65 ± 0.16 *** | 1.23 ± 0.12 # |
| HDL-C/LDL-C | 6.03 ± 0.24 | 2.33 ± 0.24 *** | 2.35 ± 0.28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Kang, S.-G.; Huang, K.; Tong, T. Dietary Supplementation of Methyl Cedryl Ether Ameliorates Adiposity in High-Fat Diet-Fed Mice. Nutrients 2023, 15, 788. https://doi.org/10.3390/nu15030788
Li M, Kang S-G, Huang K, Tong T. Dietary Supplementation of Methyl Cedryl Ether Ameliorates Adiposity in High-Fat Diet-Fed Mice. Nutrients. 2023; 15(3):788. https://doi.org/10.3390/nu15030788
Chicago/Turabian StyleLi, Mengjie, Seong-Gook Kang, Kunlun Huang, and Tao Tong. 2023. "Dietary Supplementation of Methyl Cedryl Ether Ameliorates Adiposity in High-Fat Diet-Fed Mice" Nutrients 15, no. 3: 788. https://doi.org/10.3390/nu15030788
APA StyleLi, M., Kang, S. -G., Huang, K., & Tong, T. (2023). Dietary Supplementation of Methyl Cedryl Ether Ameliorates Adiposity in High-Fat Diet-Fed Mice. Nutrients, 15(3), 788. https://doi.org/10.3390/nu15030788