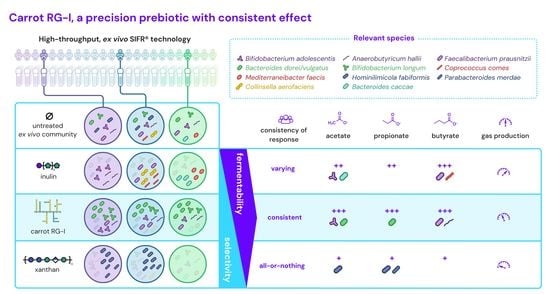
Carrot RG-I Reduces Interindividual Differences between 24 Adults through Consistent Effects on Gut Microbiota Composition and Function Ex Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Compounds
2.2. SIFR® Technology
2.3. Fundamental Fermentation Parameters
2.4. Microbiota Phylogenetic Analysis: Quantitative 16S rRNA Gene Profiling
2.5. Statistical Analysis
3. Results
3.1. The Study Cohort Covered Established Interpersonal Differences in Enterotypes Described for Human Adult Gut Microbiota
3.2. cRG-I Leads to the Most Marked and Consistent Effects on Microbiota Composition and Metabolite Production
3.3. cRG-I Was Selectively Fermented by Taxa Consistently Present in the Commensal Gut Microbiota of Human Adults across Different Enterotypes
3.4. cRG-I Most Markedly and Consistently Stimulated Acetate and Propionate Production with Only Minor Increases in Gas Production
3.5. Specific OTUs Correlated with Specific Metabolites upon cRG-I Supplementation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut Microbiota and IBD: Causation or Correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef]
- Rebersek, M. Gut Microbiome and Its Role in Colorectal Cancer. BMC Cancer 2021, 21, 1325. [Google Scholar] [CrossRef]
- Caio, G.; Lungaro, L.; Segata, N.; Guarino, M.; Zoli, G.; Volta, U.; De Giorgio, R. Effect of Gluten-Free Diet on Gut Microbiota Composition in Patients with Celiac Disease and Non-Celiac Gluten/Wheat Sensitivity. Nutrients 2020, 12, 1832. [Google Scholar] [CrossRef]
- Shen, T.; Yue, Y.; He, T.; Huang, C.; Qu, B.; Lv, W.; Lai, H.-Y. The Association Between the Gut Microbiota and Parkinson’s Disease, a Meta-Analysis. Front. Aging Neurosci. 2021, 13, 636545. [Google Scholar] [CrossRef] [PubMed]
- de Vos, W.M.; Tilg, H.; Hul, M.V.; Cani, P.D. Gut Microbiome and Health: Mechanistic Insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed]
- van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-Chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Lavelle, A.; Lennon, G.; O’Sullivan, O.; Docherty, N.; Balfe, A.; Maguire, A.; Mulcahy, H.E.; Doherty, G.; O’Donoghue, D.; Hyland, J.; et al. Spatial Variation of the Colonic Microbiota in Patients with Ulcerative Colitis and Control Volunteers. Gut 2015, 64, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.K.; Deehan, E.C.; Zhang, Z.; Jin, M.; Baskota, N.; Perez-Muñoz, M.E.; Cole, J.; Tuncil, Y.E.; Seethaler, B.; Wang, T.; et al. Gut Microbiota Modulation with Long-Chain Corn Bran Arabinoxylan in Adults with Overweight and Obesity Is Linked to an Individualized Temporal Increase in Fecal Propionate. Microbiome 2020, 8, 118. [Google Scholar] [CrossRef]
- Healey, G.R.; Murphy, R.; Brough, L.; Butts, C.A.; Coad, J. Interindividual Variability in Gut Microbiota and Host Response to Dietary Interventions. Nutr. Rev. 2017, 75, 1059–1080. [Google Scholar] [CrossRef]
- Armet, A.M.; Deehan, E.C.; Thöne, J.V.; Hewko, S.J.; Walter, J. The Effect of Isolated and Synthetic Dietary Fibers on Markers of Metabolic Diseases in Human Intervention Studies: A Systematic Review. Adv. Nutr. 2020, 11, 420–438. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the Human Gut Microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Kathagen, G.; D’hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative Microbiome Profiling Links Gut Community Variation to Microbial Load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the Landscape of Gut Microbial Community Composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-Level Analysis of Gut Microbiome Variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Wu, D.; Ye, X.; Linhardt, R.J.; Liu, X.; Zhu, K.; Yu, C.; Ding, T.; Liu, D.; He, Q.; Chen, S. Dietary Pectic Substances Enhance Gut Health by Its Polycomponent: A Review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2015–2039. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and Prebiotics in Intestinal Health and Disease: From Biology to the Clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Marasco, G.; Cirota, G.G.; Rossini, B.; Lungaro, L.; Di Biase, A.R.; Colecchia, A.; Volta, U.; De Giorgio, R.; Festi, D.; Caio, G. Probiotics, Prebiotics and Other Dietary Supplements for Gut Microbiota Modulation in Celiac Disease Patients. Nutrients 2020, 12, 2674. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Cantu-Jungles, T.M.; Hamaker, B.R. New View on Dietary Fiber Selection for Predictable Shifts in Gut Microbiota. mBio 2020, 11, e02179-19. [Google Scholar] [CrossRef] [PubMed]
- Cantu-Jungles, T.M.; Bulut, N.; Chambry, E.; Ruthes, A.; Iacomini, M.; Keshavarzian, A.; Johnson, T.A.; Hamaker, B.R. Dietary Fiber Hierarchical Specificity: The Missing Link for Predictable and Strong Shifts in Gut Bacterial Communities. mBio 2021, 12, e01028-21. [Google Scholar] [CrossRef]
- Ostrowski, M.P.; La Rosa, S.L.; Kunath, B.J.; Robertson, A.; Pereira, G.; Hagen, L.H.; Varghese, N.J.; Qiu, L.; Yao, T.; Flint, G.; et al. Mechanistic Insights into Consumption of the Food Additive Xanthan Gum by the Human Gut Microbiota. Nat. Microbiol. 2022, 7, 556–569. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Verstrepen, L.; Ghyselinck, J.; Albers, R.; Marzorati, M.; Mercenier, A. A Novel Non-Digestible, Carrot-Derived Polysaccharide (CRG-I) Selectively Modulates the Human Gut Microbiota While Promoting Gut Barrier Integrity: An Integrated In Vitro Approach. Nutrients 2020, 12, 1917. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Duysburgh, C.; Cleenwerck, I.; Albers, R.; Marzorati, M.; Mercenier, A. Consistent Prebiotic Effects of Carrot RG-I on the Gut Microbiota of Four Human Adult Donors in the SHIME® Model despite Baseline Individual Variability. Microorganisms 2021, 9, 2142. [Google Scholar] [CrossRef]
- O’Donnell, M.M.; Rea, M.C.; Shanahan, F.; Ross, R.P. The Use of a Mini-Bioreactor Fermentation System as a Reproducible, High-Throughput Ex Vivo Batch Model of the Distal Colon. Front. Microbiol. 2018, 9, 1844. [Google Scholar] [CrossRef]
- Biagini, F.; Calvigioni, M.; Lapomarda, A.; Vecchione, A.; Magliaro, C.; De Maria, C.; Montemurro, F.; Celandroni, F.; Mazzantini, D.; Mattioli-Belmonte, M.; et al. A Novel 3D in Vitro Model of the Human Gut Microbiota. Sci. Rep. 2020, 10, 21499. [Google Scholar] [CrossRef]
- Gaisawat, M.B.; MacPherson, C.W.; Tremblay, J.; Piano, A.; Iskandar, M.M.; Tompkins, T.A.; Kubow, S. Probiotic Supplementation in a Clostridium Difficile-Infected Gastrointestinal Model Is Associated with Restoring Metabolic Function of Microbiota. Microorganisms 2019, 8, E60. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-Producing Clostridium Cluster XIVa Species Specifically Colonize Mucins in an In Vitro Gut Model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; Maathuis, A.; Heilig, H.G.H.J.; Venema, K.; de Vos, W.M.; Smidt, H. Evaluating the Microbial Diversity of an in Vitro Model of the Human Large Intestine by Phylogenetic Microarray Analysis. Microbiol. (Read.) 2010, 156, 3270–3281. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Grootaert, C.; Marzorati, M.; Possemiers, S.; Verstraete, W.; Gérard, P.; Rabot, S.; Bruneau, A.; El Aidy, S.; Derrien, M.; et al. Microbial Community Development in a Dynamic Gut Model Is Reproducible, Colon Region Specific, and Selective for Bacteroidetes and Clostridium Cluster IX. Appl. Environ. Microbiol. 2010, 76, 5237–5246. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Deyaert, S.; Thabuis, C.; Perreau, C.; Bajic, D.; Wintergerst, E.; Joossens, M.; Firrman, J.; Walsh, D.; Baudot, A. Bridging Preclinical and Clinical Gut Microbiota Research Using the Ex Vivo SIFR Technology. Front. Microbiol. 2023, 14, 1043. [Google Scholar] [CrossRef]
- Hughes, J.B.; Hellmann, J.J.; Ricketts, T.H.; Bohannan, B.J.M. Counting the Uncountable: Statistical Approaches to Estimating Microbial Diversity. Appl. Environ. Microbiol. 2001, 67, 4399–4406. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLOS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal. R. Stat. Soc. Ser. B. (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Props, R.; Kerckhof, F.-M.; Rubbens, P.; De Vrieze, J.; Hernandez Sanabria, E.; Waegeman, W.; Monsieurs, P.; Hammes, F.; Boon, N. Absolute Quantification of Microbial Taxon Abundances. ISME J. 2017, 11, 584–587. [Google Scholar] [CrossRef]
- De Vuyst, L.; Moens, F.; Selak, M.; Rivière, A.; Leroy, F. Summer Meeting 2013: Growth and Physiology of Bifidobacteria. J. Appl. Microbiol. 2014, 116, 477–491. [Google Scholar] [CrossRef]
- Shetty, S.A.; Zuffa, S.; Bui, T.P.N.; Aalvink, S.; Smidt, H.; De Vos, W.M. Reclassification of Eubacterium Hallii as Anaerobutyricum Hallii Gen. Nov., Comb. Nov., and Description of Anaerobutyricum Soehngenii Sp. Nov., a Butyrate and Propionate-Producing Bacterium from Infant Faeces. Int. J. Syst. Evol. Microbiol. 2018, 68, 3741–3746. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-Utilizing Bacteria, Isolated from Human Feces, That Produce Butyrate as a Major Fermentation Product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef]
- Maturana, J.L.; Cárdenas, J.P. Insights on the Evolutionary Genomics of the Blautia Genus: Potential New Species and Genetic Content Among Lineages. Front. Microbiol. 2021, 12, 660920. [Google Scholar] [CrossRef]
- Berni Canani, R.; Sangwan, N.; Stefka, A.T.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus Rhamnosus GG-Supplemented Formula Expands Butyrate-Producing Bacterial Strains in Food Allergic Infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef]
- Duncan, S.H.; Hold, G.L.; Harmsen, H.J.M.; Stewart, C.S.; Flint, H.J. Growth Requirements and Fermentation Products of Fusobacterium prausnitzii, and a Proposal to Reclassify It as Faecalibacterium prausnitzii Gen. Nov., Comb. Nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 2141–2146. [Google Scholar] [CrossRef]
- Martens, E.C.; Lowe, E.C.; Chiang, H.; Pudlo, N.A.; Wu, M.; McNulty, N.P.; Abbott, D.W.; Henrissat, B.; Gilbert, H.J.; Bolam, D.N.; et al. Recognition and Degradation of Plant Cell Wall Polysaccharides by Two Human Gut Symbionts. PLoS Biol. 2011, 9, e1001221. [Google Scholar] [CrossRef]
- Ndeh, D.; Rogowski, A.; Cartmell, A.; Luis, A.S.; Baslé, A.; Gray, J.; Venditto, I.; Briggs, J.; Zhang, X.; Labourel, A.; et al. Complex Pectin Metabolism by Gut Bacteria Reveals Novel Catalytic Functions. Nature 2017, 544, 65–70. [Google Scholar] [CrossRef]
- O’Connell Motherway, M.; Fitzgerald, G.F.; van Sinderen, D. Metabolism of a Plant Derived Galactose-Containing Polysaccharide by Bifidobacterium Breve UCC2003. Microb. Biotechnol. 2011, 4, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Komeno, M.; Hayamizu, H.; Fujita, K.; Ashida, H. Two Novel α-l-Arabinofuranosidases from Bifidobacterium longum Subsp. longum Belonging to Glycoside Hydrolase Family 43 Cooperatively Degrade Arabinan. Appl. Environ. Microbiol. 2019, 85, e02582-18. [Google Scholar] [CrossRef]
- Kelly, S.M.; Munoz-Munoz, J.; van Sinderen, D. Plant Glycan Metabolism by Bifidobacteria. Front. Microbiol. 2021, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zheng, J.; Wang, F.; Zheng, J.; Yang, D. Dietary Fiber Chemical Structure Determined Gut Microbiota Dynamics. iMeta 2022, 1, e64. [Google Scholar] [CrossRef]
- Afrizal, A.; Hitch, T.C.A.; Viehof, A.; Treichel, N.; Riedel, T.; Abt, B.; Buhl, E.M.; Kohlheyer, D.; Overmann, J.; Clavel, T. Anaerobic Single-Cell Dispensing Facilitates the Cultivation of Human Gut Bacteria. Environ. Microbiol. 2022, 24, 3861–3881. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Wang, J.; Sailer, M.; Theis, S.; Verbeke, K.; Raes, J. Prebiotic Inulin-Type Fructans Induce Specific Changes in the Human Gut Microbiota. Gut 2017, 66, 1968–1974. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Gérard, P.; Rabot, S.; Bruneau, A.; El Aidy, S.; Derrien, M.; Kleerebezem, M.; Zoetendal, E.G.; Smidt, H.; Verstraete, W.; et al. Arabinoxylans and Inulin Differentially Modulate the Mucosal and Luminal Gut Microbiota and Mucin-Degradation in Humanized Rats. Environ. Microbiol. 2011, 13, 2667–2680. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Thomson, J.M.; Flint, H.J. The Role of PH in Determining the Species Composition of the Human Colonic Microbiota. Environ. Microbiol. 2009, 11, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Lin, Y.; Zhang, H.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life. Nutrients 2022, 14, 1498. [Google Scholar] [CrossRef]
- Derrien, M.; Turroni, F.; Ventura, M.; van Sinderen, D. Insights into Endogenous Bifidobacterium Species in the Human Gut Microbiota during Adulthood. Trends Microbiol. 2022, 30, 940–947. [Google Scholar] [CrossRef]
- Alessandri, G.; Ossiprandi, M.C.; MacSharry, J.; van Sinderen, D.; Ventura, M. Bifidobacterial Dialogue with Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 2019, 10, 2348. [Google Scholar] [CrossRef]
- Wong, C.B.; Odamaki, T.; Xiao, J.-Z. Insights into the Reason of Human-Residential Bifidobacteria (HRB) Being the Natural Inhabitants of the Human Gut and Their Potential Health-Promoting Benefits. FEMS Microbiol. Rev. 2020, 44, 369–385. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent Development of Probiotic Bifidobacteria for Treating Human Diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium Prausnitzii Is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. PNAS 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Almeida, D.; Machado, D.; Andrade, J.C.; Mendo, S.; Gomes, A.M.; Freitas, A.C. Evolving Trends in Next-Generation Probiotics: A 5W1H Perspective. Crit. Rev. Food Sci. Nutr. 2020, 60, 1783–1796. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Emoto, T.; Yamashita, T.; Watanabe, H.; Hayashi, T.; Tabata, T.; Hoshi, N.; Hatano, N.; Ozawa, G.; Sasaki, N.; et al. Bacteroides Vulgatus and Bacteroides Dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis. Circulation 2018, 138, 2486–2498. [Google Scholar] [CrossRef] [PubMed]
- Zafar, H.; Saier, M.H. Gut Bacteroides Species in Health and Disease. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Song, L.; Huang, Y.; Liu, G.; Li, X.; Xiao, Y.; Liu, C.; Zhang, Y.; Li, J.; Xu, J.; Lu, S.; et al. A Novel Immunobiotics Bacteroides Dorei Ameliorates Influenza Virus Infection in Mice. Front. Immunol. 2021, 12, 828887. [Google Scholar] [CrossRef]
- Livesey, G. Tolerance of Low-Digestible Carbohydrates: A General View. Br. J. Nutr. 2001, 85, S7–S16. [Google Scholar] [CrossRef] [PubMed]
- Marteau, P.; Seksik, P. Tolerance of Probiotics and Prebiotics. J. Clin. Gastroenterol. 2004, 38, S67–S69. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van den Abbeele, P.; Deyaert, S.; Albers, R.; Baudot, A.; Mercenier, A. Carrot RG-I Reduces Interindividual Differences between 24 Adults through Consistent Effects on Gut Microbiota Composition and Function Ex Vivo. Nutrients 2023, 15, 2090. https://doi.org/10.3390/nu15092090
Van den Abbeele P, Deyaert S, Albers R, Baudot A, Mercenier A. Carrot RG-I Reduces Interindividual Differences between 24 Adults through Consistent Effects on Gut Microbiota Composition and Function Ex Vivo. Nutrients. 2023; 15(9):2090. https://doi.org/10.3390/nu15092090
Chicago/Turabian StyleVan den Abbeele, Pieter, Stef Deyaert, Ruud Albers, Aurélien Baudot, and Annick Mercenier. 2023. "Carrot RG-I Reduces Interindividual Differences between 24 Adults through Consistent Effects on Gut Microbiota Composition and Function Ex Vivo" Nutrients 15, no. 9: 2090. https://doi.org/10.3390/nu15092090
APA StyleVan den Abbeele, P., Deyaert, S., Albers, R., Baudot, A., & Mercenier, A. (2023). Carrot RG-I Reduces Interindividual Differences between 24 Adults through Consistent Effects on Gut Microbiota Composition and Function Ex Vivo. Nutrients, 15(9), 2090. https://doi.org/10.3390/nu15092090