Anti-Inflammatory Mechanisms of Curcumin and Its Metabolites in White Adipose Tissue and Cultured Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Studies
2.2. RNA Extraction, cDNA Preparation, and Quantitative Real-Time PCR
2.3. Measurement of THC and COG in Curcumin, Serum, and Adipocytes
2.4. Cell Culture Studies
2.5. 3T3-L1 Cell Culture Treatments
2.6. Oil Red O Staining (Lipid Staining) of Curcumin-Treated Adipocytes
2.7. ELISA Cytokine Assays
2.8. Protein Analysis by Mass Spectrometry
2.8.1. Protein Extraction, Tryptic Digestion, and Analysis by LC-MS/MS
2.8.2. Protein Identification, Label-Free Quantification, and Data Analyses
2.8.3. Pathway and Network Analyses
2.9. Statistical Analyses
3. Results
3.1. Mouse Data
Curcumin Metabolites Tetrahydrocurcumin (THC) and Curcumin-O-Glucuronide (COG) in Serum
3.2. Adipose Cell Culture
3.2.1. Cell Viability and Dose–Response Studies
3.2.2. Effect of Curcumin on Lipid Accumulation during Adipocyte Differentiation
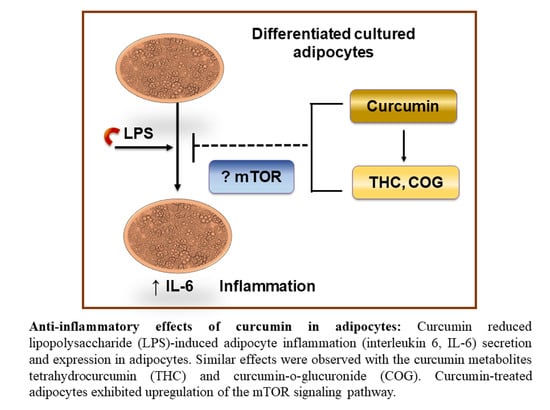
3.2.3. Curcumin Metabolites THC and COG Reduced IL-6 Secretion in LPS (200 ng/mL)-Induced Differentiated 3T3-L1 Adipocytes
3.2.4. Detection of THC and COG in Curcumin-Treated Adipocytes
3.2.5. Proteomic Analyses of Curcumin-Treated Adipocytes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| ABC | Ammonium bicarbonate | mTOR | Mammalian target of rapamycin |
| Ampk | AMP-activated protein kinase | NfkB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| COG | Curcumin-o-glucuronide | LPS | Lipopolysaccharide |
| DIO | Diet-induced obesity | LBP | LPS-binding protein |
| eIF2 | Eukaryotic translation initiation factor 2 | NRF1 | Nuclear respiratory factor 1 |
| FFA | Free fatty acid | Ppar-γ | Peroxisome proliferator-activated receptor gamma |
| HFD | High-fat diet | Stat1 | Signal transducer and activator of transcription 1 |
| HFC | High-fat diet with curcumin | Sirt1 | Sirtuin 1 |
| IkB | Inhibitory kappa-B | ||
| IL-6 | Interleukin-6 | THC | Tetrahydrocurcumin |
| IPA | Ingenuity pathway analysis | Tlr4 | Toll-like receptor-4 |
| IRF3 | interferon regulatory factor 3 | TMP | Transmembrane proteins |
| Irs1 | Insulin receptor substrate 1 | T2D | Type 2 diabetes |
| ISR | Integrated stress response | TNF-α | Tumor necrosis factor-alpha |
| LFQ | Label-free quantification | TRIF | Toll/IL-1R domain-containing adaptor-inducing IFN-β |
| Mcp1 | Monocyte chemoattractant protein-1 | WAT | White adipose tissue |
References
- Ren, Y.; Zhao, H.; Yin, C.; Lan, X.; Wu, L.; Du, X.; Griffiths, H.R.; Gao, D. Adipokines, Hepatokines and Myokines: Focus on Their Role and Molecular Mechanisms in Adipose Tissue Inflammation. Front. Endocrinol. 2022, 13, 873699. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Koboziev, I.; Albracht-Schulte, K.; Mistretta, B.; Scoggin, S.; Yosofvand, M.; Moussa, H.; Zabet-Moghaddam, M.; Ramalingam, L.; Gunaratne, P.H.; et al. Curcumin Reduces Adipose Tissue Inflammation and Alters Gut Microbiota in Diet-Induced Obese Male Mice. Mol. Nutr. Food Res. 2021, 65, 2100274. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, E.A.; Trogdon, J.G.; Cohen, J.W.; Dietz, W.H. Annual medical spending attributable to obesity: Payer-and service-specific estimates. Health Aff. (Millwood) 2009, 28, w822–w831. [Google Scholar] [CrossRef] [PubMed]
- Withrow, D.; Alter, D.A. The economic burden of obesity worldwide: A systematic review of the direct costs of obesity. Obes. Rev. 2011, 12, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Jain, P.; Hassan, N.; Farooq, U.; Mirza, M.A.; Pandith, A.A.; Iqbal, Z. Role of Genetic and Dietary Implications in the Pathogenesis of Global Obesity. Food Rev. Int. 2021, 38, 434–455. [Google Scholar] [CrossRef]
- Theilade, S.; Christensen, M.B.; Vilsbøll, T.; Knop, F.K. An overview of obesity mechanisms in humans: Endocrine regulation of food intake, eating behaviour and common determinants of body weight. Diabetes Obes. Metab. 2021, 23, 17–35. [Google Scholar] [CrossRef] [PubMed]
- D’Antongiovanni, V.; Fornai, M.; Pellegrini, C.; Blandizzi, C.; Antonioli, L. Managing Obesity and Related Comorbidities: A Potential Pharmacological Target in the Adenosine System? Front. Pharmacol. 2021, 11, 621955. [Google Scholar] [CrossRef]
- Ahmad, F.B.; Anderson, R.N. The Leading Causes of Death in the US for 2020. JAMA 2021, 325, 1829–1830. [Google Scholar] [CrossRef]
- Siiteri, P.K. Adipose tissue as a source of hormones. Am. J. Clin. Nutr. 1987, 45, 277–282. [Google Scholar] [CrossRef]
- Frayn, K.N.; Karpe, F.; Fielding, B.A.; Macdonald, I.A.; Coppack, S.W. Integrative physiology of human adipose tissue. Int. J. Obes. Relat. Metab. Disord. 2003, 27, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Kalupahana, N.S.; Goonapienuwala, B.L.; Moustaid-Moussa, N. Omega-3 Fatty Acids and Adipose Tissue: Inflammation and Browning. Annu. Rev. Nutr. 2020, 40, 25–49. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K. Fat uses a TOLL-road to connect inflammation and diabetes. Cell Metab. 2006, 4, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Rohm, T.V.; Meier, D.T.; Olefsky, J.M.; Donath, M.Y. Inflammation in obesity, diabetes, and related disorders. Immunity 2022, 55, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Kahn, B.B.; Flier, J.S. Obesity and insulin resistance. J. Clin. Investig. 2000, 106, 473–481. [Google Scholar] [CrossRef]
- Kim, J.-E.; Kim, J.-S.; Jo, M.-J.; Cho, E.; Ahn, S.-Y.; Kwon, Y.-J.; Ko, G.-J. The Roles and Associated Mechanisms of Adipokines in Development of Metabolic Syndrome. Molecules 2022, 27, 334. [Google Scholar] [CrossRef]
- Alkhatib, D.H.; Jaleel, A.; Tariq, M.N.M.; Feehan, J.; Apostolopoulos, V.; Ismail, L.C.; Stojanovska, L.; Al Dhaheri, A.S. The Role of Bioactive Compounds from Dietary Spices in the Management of Metabolic Syndrome: An Overview. Nutrients 2022, 14, 175. [Google Scholar] [CrossRef]
- Miao, L.; Huang, F.; Sun, Y.-Y.; Jiang, W.; Chen, Y.-J.; Zhang, M. Curcumin plays a local anti-inflammatory and antioxidant role via the HMGB1/TLR4/NF-ΚB pathway in rat masseter muscle under psychological stress. J. Oral Rehabil. 2022, 49, 249–257. [Google Scholar] [CrossRef]
- Carpene, C.; Gomez-Zorita, S.; Deleruyelle, S.; Carpene, M. Novel strategies for preventing diabetes and obesity complications with natural polyphenols. Curr. Med. Chem. 2015, 22, 150–164. [Google Scholar] [CrossRef]
- Guo, X.; Tresserra-Rimbau, A.; Estruch, R.; Martínez-González, M.A.; Medina-Remón, A.; Fitó, M.; Corella, D.; Salas-Salvadó, J.; Portillo, M.P.; Moreno, J.J.; et al. Polyphenol Levels Are Inversely Correlated with Body Weight and Obesity in an Elderly Population after 5 Years of Follow Up (The Randomised PREDIMED Study). Nutrients 2017, 9, 452. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Zhu, R.; Liu, Q.; Fei, J.; Wang, S. Anti-inflammatory activity of curcumin-loaded solid lipid nanoparticles in IL-1beta transgenic mice subjected to the lipopolysaccharide-induced sepsis. Biomaterials 2015, 53, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.S.; He, H.; Wang, J.; Gehr, T.W.; Ghosh, S. Curcumin-mediated regulation of intestinal barrier function: The mechanism underlying its beneficial effects. Tissue Barriers 2018, 6, e1425085. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.-H.; Chen, J.-W.; Kong, Z.-L.; Wu, J.-C.; Ho, C.-T.; Lai, C.-S. Attenuation by Tetrahydrocurcumin of Adiposity and Hepatic Steatosis in Mice with High-Fat-Diet-Induced Obesity. J. Agric. Food Chem. 2018, 66, 12685–12695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-B.; Luo, D.-D.; Xie, J.-H.; Xian, Y.-F.; Lai, Z.-Q.; Liu, Y.-H.; Liu, W.-H.; Chen, J.-N.; Lai, X.-P.; Lin, Z.-X.; et al. Curcumin’s Metabolites, Tetrahydrocurcumin and Octahydrocurcumin, Possess Superior Anti-inflammatory Effects in vivo Through Suppression of TAK1-NF-κB Pathway. Front. Pharmacol. 2018, 9, 1181. [Google Scholar] [CrossRef] [PubMed]
- Jayarathne, S.; Stull, A.J.; Miranda, A.; Scoggin, S.; Claycombe-Larson, K.; Kim, J.H.; Moustaid-Moussa, N. Tart Cherry Reduces Inflammation in Adipose Tissue of Zucker Fatty Rats and Cultured 3T3-L1 Adipocytes. Nutrients 2018, 10, 1576. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [PubMed]
- Kobori, M.; Takahashi, Y.; Takeda, H.; Takahashi, M.; Izumi, Y.; Akimoto, Y.; Sakurai, M.; Oike, H.; Nakagawa, T.; Itoh, M.; et al. Dietary Intake of Curcumin Improves eIF2 Signaling and Reduces Lipid Levels in the White Adipose Tissue of Obese Mice. Sci. Rep. 2018, 8, 9081. [Google Scholar] [CrossRef] [PubMed]
- Khalid, M.; Alkaabi, J.; Khan, M.A.B.; Adem, A. Insulin Signal Transduction Perturbations in Insulin Resistance. Int. J. Mol. Sci. 2021, 22, 8590. [Google Scholar] [CrossRef]
- Um, S.H.; Frigerio, F.; Watanabe, M.; Picard, F.; Joaquin, M.; Sticker, M.; Fumagalli, S.; Allegrini, P.R.; Kozma, S.C.; Auwerx, J.; et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004, 431, 200–205. [Google Scholar] [CrossRef]
- Kanda, H.; Tateya, S.; Tamori, Y.; Kotani, K.; Hiasa, K.-I.; Kitazawa, R.; Kitazawa, S.; Miyachi, H.; Maeda, S.; Egashira, K.; et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Investig. 2006, 116, 1494–1505. [Google Scholar] [CrossRef]
- Ahmad, R.; Al-Roub, A.; Kochumon, S.; Akther, N.; Thomas, R.; Kumari, M.; Koshy, M.S.; Tiss, A.; Hannun, Y.A.; Tuomilehto, J.; et al. The Synergy between Palmitate and TNF-α for CCL2 Production Is Dependent on the TRIF/IRF3 Pathway: Implications for Metabolic Inflammation. J. Immunol. 2018, 200, 3599–3611. [Google Scholar] [CrossRef] [PubMed]
- Park, M.-Y.; Mun, S.T. Carnosic acid inhibits TLR4-MyD88 signaling pathway in LPS-stimulated 3T3-L1 adipocytes. Nutr. Res. Pract. 2014, 8, 516. [Google Scholar] [CrossRef] [PubMed]
- Meijer, K.; de Vries, M.; Al-Lahham, S.; Bruinenberg, M.; Weening, D.; Dijkstra, M.; Kloosterhuis, N.; van der Leij, R.J.; van der Want, H.; Kroesen, B.-J.; et al. Human Primary Adipocytes Exhibit Immune Cell Function: Adipocytes Prime Inflammation Independent of Macrophages. PLoS ONE 2011, 6, e17154. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.-M.; Kang, J.-H.; Kawada, T.; Yoo, H.; Sung, M.-K.; Yu, R. Active spice-derived components can inhibit inflammatory responses of adipose tissue in obesity by suppressing inflammatory actions of macrophages and release of monocyte chemoattractant protein-1 from adipocytes. Life Sci. 2007, 80, 926–931. [Google Scholar] [CrossRef]
- Ciardi, C.; Jenny, M.; Tschoner, A.; Ueberall, F.; Patsch, J.; Pedrini, M.; Ebenbichler, C.; Fuchs, D. Food additives such as sodium sulphite, sodium benzoate and curcumin inhibit leptin release in lipopolysaccharide-treated murine adipocytes in vitro. Br. J. Nutr. 2012, 107, 826–833. [Google Scholar] [CrossRef]
- Wang, S.-L.; Li, Y.; Wen, Y.; Chen, Y.-F.; Na, L.-X.; Li, S.-T.; Sun, C.-H. Curcumin, a Potential Inhibitor of Up-regulation of TNF-alpha and IL-6 Induced by Palmitate in 3T3-L1 Adipocytes through NF-kappaB and JNK Pathway. Biomed. Environ. Sci. 2009, 22, 32–39. [Google Scholar] [CrossRef]
- Varì, R.; Scazzocchio, B.; Silenzi, A.; Giovannini, C.; Masella, R. Obesity-Associated Inflammation: Does Curcumin Exert a Beneficial Role? Nutrients 2021, 13, 1021. [Google Scholar] [CrossRef]
- Oyadomari, S.; Harding, H.P.; Zhang, Y.; Oyadomari, M.; Ron, D. Dephosphorylation of Translation Initiation Factor 2α Enhances Glucose Tolerance and Attenuates Hepatosteatosis in Mice. Cell Metab. 2008, 7, 520–532. [Google Scholar] [CrossRef]
- Marton, L.T.; Pescinini-E-Salzedas, L.M.; Camargo, M.E.C.; Barbalho, S.M.; Haber, J.F.d.S.; Sinatora, R.V.; Detregiachi, C.R.P.; Girio, R.J.S.; Buchaim, D.V.; Bueno, P.C.d.S. The Effects of Curcumin on Diabetes Mellitus: A Systematic Review. Front. Endocrinol. 2021, 12, 669448. [Google Scholar] [CrossRef]
- Casimiro, I.; Stull, N.D.; Tersey, S.A.; Mirmira, R.G. Phenotypic sexual dimorphism in response to dietary fat manipulation in C57BL/6J mice. J. Diabetes Complicat. 2021, 35, 107795. [Google Scholar] [CrossRef]
- Lund, N.C.; Kayode, Y.; McReynolds, M.R.; Clemmer, D.C.; Hudson, H.; Clerc, I.; Hong, H.-K.; Brenchley, J.M.; Bass, J.; D’aquila, R.T.; et al. mTOR regulation of metabolism limits LPS-induced monocyte inflammatory and procoagulant responses. Commun. Biol. 2022, 5, 878. [Google Scholar] [CrossRef] [PubMed]





| Samples | THC Detection | COG Detection |
|---|---|---|
| Control (no LPS or curcumin) | No | No |
| LPS + curcumin-treated adipocytes | Yes | Not analyzed |
| Curcumin powder | No | Yes |
| Ingenuity Canonical Pathways | −log (p-Value) | Ratio | z-Score |
|---|---|---|---|
| EIF2 Signaling | 15.2 | 0.0938 | 3 |
| Regulation of eIF4 and p70S6K Signaling | 10.3 | 0.0838 | - |
| mTOR Signaling | 9.25 | 0.0708 | 1.342 |
| BAG2 Signaling Pathway | 8.52 | 0.119 | - |
| FAT10 Signaling Pathway | 7.56 | 0.143 | - |
| Glutathione Redox Reactions II | 5.6 | 0.75 | - |
| Protein Ubiquitination Pathway | 5.27 | 0.0436 | - |
| Sirtuin Signaling Pathway | 4.29 | 0.0377 | 0.816 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, T.; Scoggin, S.; Gong, X.; Zabet-Moghaddam, M.; Kalupahana, N.S.; Moustaid-Moussa, N. Anti-Inflammatory Mechanisms of Curcumin and Its Metabolites in White Adipose Tissue and Cultured Adipocytes. Nutrients 2024, 16, 70. https://doi.org/10.3390/nu16010070
Islam T, Scoggin S, Gong X, Zabet-Moghaddam M, Kalupahana NS, Moustaid-Moussa N. Anti-Inflammatory Mechanisms of Curcumin and Its Metabolites in White Adipose Tissue and Cultured Adipocytes. Nutrients. 2024; 16(1):70. https://doi.org/10.3390/nu16010070
Chicago/Turabian StyleIslam, Tariful, Shane Scoggin, Xiaoxia Gong, Masoud Zabet-Moghaddam, Nishan S. Kalupahana, and Naima Moustaid-Moussa. 2024. "Anti-Inflammatory Mechanisms of Curcumin and Its Metabolites in White Adipose Tissue and Cultured Adipocytes" Nutrients 16, no. 1: 70. https://doi.org/10.3390/nu16010070
APA StyleIslam, T., Scoggin, S., Gong, X., Zabet-Moghaddam, M., Kalupahana, N. S., & Moustaid-Moussa, N. (2024). Anti-Inflammatory Mechanisms of Curcumin and Its Metabolites in White Adipose Tissue and Cultured Adipocytes. Nutrients, 16(1), 70. https://doi.org/10.3390/nu16010070