Comparative Proteomics of Peritrophic Matrix Provides an Insight into its Role in Cry1Ac Resistance of Cotton Bollworm Helicoverpa armigera
Abstract
:1. Introduction
2. Results
2.1. Resistance to Cry1Ac Protoxin and Activated Toxin
2.2. Transcriptomic Analysis of Midguts from XJ and XJ10 Larvae
2.3. Proteomic Analysis of PM from XJ and XJ10 Larvae
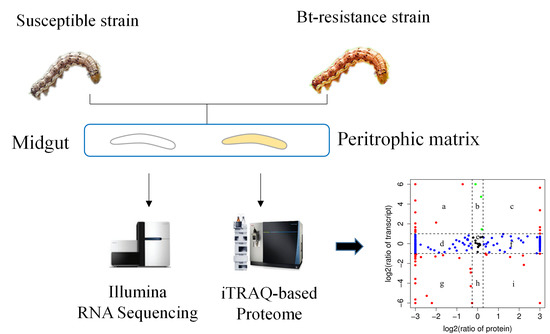
2.4. Correlation between Transcriptome and Proteome
3. Discussion
3.1. Identification of Potential Resistance Mechanisms in XJ10
3.2. Identification of PM Proteins
4. Materials and Methods
4.1. Insect Strains
4.2. Insect Bioassays
4.3. RNA Extraction and cDNA Library Construction
4.4. Data Processing and Analysis
4.5. qRT-PCR Analysis of Gene Expression Levels
4.6. iTRAQ-Based Proteome Determination and Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mendelsohn, M.; Kough, J.; Vaituzis, Z.; Matthews, K. Are bt crops safe? Nat. Biotechnol. 2003, 21, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Liliana, P.L.; Mario, S.; Alejandra, B. Bacillus thuringiensis insecticidal three-domain cry toxins: Mode of action, insect resistance and consequences for crop protection. Fems Microbiol. Rev. 2013, 37, 3–22. [Google Scholar]
- Van Frankenhuyzen, K. Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 2009, 101, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hilbeck, A.; Otto, M. Specificity and combinatorial effects of Bacillus thuringiensis cry toxins in the context of GMO environmental risk assessment. Front. Environ. Sci. 2015, 3, 71. [Google Scholar] [CrossRef]
- Wu, K.M.; Guo, Y.Y. The evolution of cotton pest management practices in china. Annu. Rev. Entomol. 2005, 50, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Kong-Ming, W.; Yan-Hui, L.; Hong-Qiang, F.; Yu-Ying, J.; Jian-Zhou, Z. Suppression of cotton bollworm in multiple crops in china in areas with Bt toxin-containing cotton. China Basic Sci. 2008, 321, 1676–1678. [Google Scholar]
- Raman, R.; Naresh, A.; Swaminathan, S.; Rao, N.G.V.; Nimbalkar, S.A.; Bhatnagar, R.K. Resistance of Helicoverpa armigera to Cry1Ac toxin from Bacillus thuringiensis is due to improper processing of the protoxin. Biochem. J. 2009, 419, 309–316. [Google Scholar]
- Tabashnik, B.E. Evolution of resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 1994, 39, 47–79. [Google Scholar] [CrossRef]
- Tabashnik, B.E.; Huang, F.; Ghimire, M.N.; Leonard, B.R.; Siegfried, B.D.; Rangasamy, M.; Yang, Y.; Wu, Y.; Gahan, L.J.; Heckel, D.G.; et al. Efficacy of genetically modified Bt toxins against insects with different genetic mechanisms of resistance. Nat. Biotechnol. 2011, 29, 1128–1131. [Google Scholar] [CrossRef] [Green Version]
- Tabashnik, B.E.; Brévault, T.; Carrière, Y. Insect resistance to Bt crops: Lessons from the first billion acres. Nat. Biotechnol. 2013, 31, 510–521. [Google Scholar] [CrossRef]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S.; Soberon, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Cowles, E.A.; Pietrantonio, P.V. The mode of action of Bacillus thuringiensis endotoxins. Annu. Rev. Entomol. 1992, 37, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Adang, M.J.; Crickmore, N.; Jurat-Fuentes, J.L. Chapter two–diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Adv. Insect Physiol. 2014, 47, 39–87. [Google Scholar]
- Liu, C.; Xiao, Y.; Li, X.; Oppert, B.; Tabashnik, B.E.; Wu, K. Cis-mediated down-regulation of a trypsin gene associated with Bt resistance in cotton bollworm. Sci. Rep. 2014, 4, 7219. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Zhang, M.; Fabrick, J.A.; Wu, Y.; Gao, M.; Huang, F.; Wei, J.; Zhang, J.; Yelich, A.; Unnithan, G.C. Dual mode of action of Bt proteins: Protoxin efficacy against resistant insects. Sci. Rep. 2015, 5, 15107. [Google Scholar] [CrossRef]
- Oppert, B.; Kramer, K.J.; Beeman, R.W.; Johnson, D.; Mcgaughey, W.H. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J. Biol. Chem. 1997, 272, 23473–23476. [Google Scholar] [CrossRef]
- Wu, Y. Detection and mechanisms of resistance evolved in insects to cry toxins from Bacillus thuringiensis. Adv. Insect Physiol. 2014, 47, 297–342. [Google Scholar]
- Wei, J.; Liang, G.; Wang, B.; Zhong, F.; Chen, L.; Khaing, M.M.; Zhang, J.; Guo, Y.; Wu, K.; Tabashnik, B.E. Activation of Bt protoxin Cry1Ac in resistant and susceptible cotton bollworm. PLoS ONE 2016, 11, e0156560. [Google Scholar] [CrossRef]
- Luo, S.; Wang, G.; Liang, G.; Wu, K.M.; Bai, L.; Ren, X.; Guo, Y. Binding of three Cry1a toxins in resistant and susceptible strains of cotton bollworm (Helicoverpa armigera). Pestic. Biochem. Phys. 2006, 85, 104–109. [Google Scholar] [CrossRef]
- Kaur, P.; Dilawari, V.K. Inheritance of resistance to Bacillus thuringiensis Cry1Ac toxin in Helicoverpa armigera (hübner) (lepidoptera: Noctuidae) from India. Pest Manag. Sci. 2011, 67, 1294–1302. [Google Scholar] [CrossRef]
- Xinjun, X.; Liangying, Y.; Yidong, W. Disruption of a cadherin gene associated with resistance to Cry1Ac {delta}-endotoxin of bacillus thuringiensis in Helicoverpa armigera. Appl. Environ. Microbiol. 2005, 71, 948–954. [Google Scholar]
- Josephakhurst, R.; Janebird, L.; Beard, C. Resistance to the Cry1Ac δ-endotoxin of Bacillus thuringiensis in the cotton bollworm, Helicoverpa armigera (lepidoptera: Noctuidae). J. Econ. Entomol. 2003, 96, 1290–1299. [Google Scholar]
- Xiao, Y.; Zhang, T.; Liu, C.; Heckel, D.G.; Li, X.; Tabashnik, B.E.; Wu, K. Mis-splicing of the abcc2 gene linked with bt toxin resistance in Helicoverpa armigera. Sci. Rep. 2014, 4, 6184. [Google Scholar] [CrossRef] [PubMed]
- Schnepf, E.; Crickmore, N.; Van Rie, J.; Lereclus, D.; Baum, J.; Feitelson, J.; Zeigler, D.R.; Dean, D.H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 775–806. [Google Scholar] [PubMed]
- Heckel, D.G.; Gahan, L.J.; Baxter, S.W.; Zhao, J.Z.; Shelton, A.M.; Gould, F.; Tabashnik, B.E. The diversity of bt resistance genes in species of lepidoptera. J. Invertebr. Pathol. 2007, 95, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Terra, W.R. The origin and functions of the insect peritrophic membrane and peritrophic gel. Arch. Insect Biochem. 2010, 47, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, D.; Erlandson, M.; Gillott, C.; Toprak, U. New insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 2009, 54, 285. [Google Scholar] [CrossRef]
- Toprak, U.; Hegedus, D.D.; Baldwin, D.; Coutu, C.; Erlandson, M. Spatial and temporal synthesis of Mamestra configurata peritrophic matrix through a larval stadium. Insect Biochem. Mol. Biol. 2014, 54, 89–97. [Google Scholar] [CrossRef]
- Campbell, P.M.; Cao, A.T.; Hines, E.R.; East, P.D.; Gordon, K.H. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem. Mol. Biol. 2008, 38, 950–958. [Google Scholar] [CrossRef]
- Wang, P.; Granados, R.R. Molecular structure of the peritrophic membrane (pm): Identification of potential PM target sites for insect control. Arch. Insect Biochem. 2010, 47, 110–118. [Google Scholar] [CrossRef]
- Wang, P.; Granados, R.R. Calcofluor disrupts the midgut defense system in insects. Insect Biochem. Mol. Biol. 2000, 30, 135–143. [Google Scholar] [CrossRef]
- Tibor, P.; Allen, C.; Paul, W.; Dawn, S.L. Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proc. Natl. Acad. Sci. USA 2002, 99, 13319–13323. [Google Scholar] [Green Version]
- Cao, G.; Zhang, L.; Liang, G.; Li, X.; Wu, K. Involvement of nonbinding site proteinases in the development of resistance of Helicoverpa armigera (Lepidoptera: Noctuidae) to Cry1Ac. J. Econ. Entomol. 2013, 106, 2514–2521. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiu, J.F.; Cheng, J.Z.; Luo, M.; Zhao, P.; Shang, X.L.; Wang, T.; Jian Wei, W.U. Proteomic analysis of the peritrophic matrix from the midgut of third instar larvae, Musca domestica. Biomed. Environ. Sci. 2016, 29, 56–65. [Google Scholar] [PubMed]
- Wang, L.; Li, F.; Wang, B.; Xiang, J. Structure and partial protein profiles of the peritrophic membrane (PM) from the gut of the shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2012, 33, 1285–1291. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Sneh, B.; Strizhov, N.; Prudovsky, E.; Regev, A.; Koncz, C.; Schell, J.; Zilberstein, A. Digestion of delta-endotoxin by gut proteases may explain reduced sensitivity of advanced instar larvae of Spodoptera littoralis to CryIc. Insect Biochem. Mol. Biol. 1996, 26, 365–373. [Google Scholar] [CrossRef]
- Karumbaiah, L.; Oppert, B.; Jurat-Fuentes, J.L.; Adang, M.J. Analysis of midgut proteinases from Bacillus thuringiensis -susceptible and -resistant Heliothis virescens (lepidoptera: Noctuidae). Comp. Biochem. Phys. B 2007, 146, 139–146. [Google Scholar] [CrossRef]
- Li, H.; Oppert, B.; Higgins, R.A.; Huang, F.; Zhu, K.Y.; Buschman, L.L. Comparative analysis of proteinase activities of Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis (Lepidoptera:Crambidae). Insect Biochem. Mol. Biol. 2004, 34, 753–762. [Google Scholar] [CrossRef]
- Gunning, R.V.; Dang, H.T.; Kemp, F.C.; Nicholson, I.C.; Moores, G.D. New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl. Environ. Microbiol. 2005, 71, 2558–2563. [Google Scholar] [CrossRef]
- Juan Luis, J.F.; Gould, F.L.; Adang, M.J. Altered glycosylation of 63- and 68-kilodalton microvillar proteins in Heliothis virescens correlates with reduced Cry1 toxin binding, decreased pore formation, and increased resistance to Bacillus thuringiensis Cry1 toxins. Appl. Environ. Microbiol. 2002, 68, 5711–5717. [Google Scholar]
- Zhang, S.; Cheng, H.; Gao, Y.; Wang, G.; Liang, G.; Wu, K. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 2009, 39, 421–429. [Google Scholar] [CrossRef]
- Herrero, S.; Gechev, T.; Bakker, P.L.; Moar, W.J.; Maagd, R.A.D. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four aminopeptidase N genes. BMC Genom. 2005, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Tiewsiri, K.; Wang, P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. USA 2011, 108, 14037–14042. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, S.; Bätz, T.; Armbruster, K.; Krasnow, M.A. Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 2006, 16, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Jacobs-Lorena, M. Characterization of a novel gut-specific chitinase gene from the human malaria vector Anopheles gambiae. J. Biol. Chem. 1997, 272, 28895–28900. [Google Scholar] [CrossRef] [PubMed]
- Ewelina, C.; Wojciech, B.A.; Monika, B.; Wlodzimierz, G. Cloning, expression, and purification of insect (Sitophilus oryzae) alpha-amylase, able to digest granular starch, in Yarrowia lipolytica host. Appl. Microbiol. Biotechnol. 2015, 99, 2727–2739. [Google Scholar]
- Gorman, M.J.; Paskewitz, S.M. Serine proteases as mediators of mosquito immune responses. Insect Biochem. Mol. Biol. 2001, 31, 257–262. [Google Scholar] [CrossRef]
- Wolfson, J.L.; Murdock, L.L. Diversity in digestive proteinase activity among insects. J. Chem. Ecol. 1990, 16, 1089–1102. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14. [Google Scholar] [CrossRef]
- Li, B. Rsem: Accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef]




| Strain | Form of Cry1Ac | LC50 a (95% Confidence Limits) | Resistance Ratio b |
|---|---|---|---|
| XJ | protoxin | 8.41 (6.31–11.25) | 1 |
| XJ10 | protoxin | 1233.91 (923.58–1665.62) | 146.79 |
| XJ | activated toxin | 7.16 (5.25–9.76) | 1 |
| XJ10 | activated toxin | 322.48 (248.42–417.66) | 45.05 |
| Gene ID | FC a | Padj Value | Description | Mechanisms |
|---|---|---|---|---|
| XM_021337885.1 | 2.53 | 1.44 × 10−6 | trypsin | Altered activation of Cry toxins |
| XM_021345079.1 | 1.54 | 3.60 × 10−17 | trypsin-like protease | |
| XM_021337887.1 | −2.69 | 7.99 × 10−10 | trypsin | |
| XM_021340602.1 | −3.56 | 5.44 × 10−9 | trypsin T4 | |
| XM_021340597.1 | −4.29 | 3.58 × 10−26 | trypsin | |
| XM_021342117.1 | −4.59 | 6.80 × 10−11 | trypsin-like proteinase T2α | |
| XM_021340588.1 | −8.89 | 0.000297197 | trypsin | |
| XM_021345839.1 | 2.41 | 6.13 × 10−24 | chymotrypsin | |
| XM_021345877.1 | 1.96 | 1.30 × 10−11 | chymotrypsin-like protease | |
| XM_021334546.1 | −5.27 | 3.40 × 10−24 | chymotrypsin | |
| XM_021345783.1 | −5.53 | 3.39 × 10−67 | chymotrypsin-like protease | |
| XM_021337546.1 | 2.03 | 1.97 × 10−5 | esterase E4-like | Sequestering the toxin |
| XM_021334360.1 | −1.70 | 5.59 × 10−10 | ABC transporter G family 23 | ABCs |
| XM_021345614.1 | 2.02 | 8.88 × 10−19 | ABCC1 protein | |
| XM_021334935.1 | −1.81 | 1.17 × 10−11 | Polycalin | Binding proteins |
| XM_021337081.1 | 1.66 | 1.17 × 10−10 | aminopeptidase N1 | |
| XM_021339316.1 | 2.24 | 6.29 × 10−11 | alkaline phosphatase 2 |
| Name | Number of Distinct Peptides | MS/MS Number | Sequence Coverage (%) | Predicted MW (kDa) | Accession Number |
|---|---|---|---|---|---|
| Chitin associated | |||||
| mucin 17-like | 1 | 6 | 0.3 | 639.67 | XM_021332791.1 |
| chitin deacetylase 5a | 3 | 12 | 7.4 | 45.297 | XM_021341184.1 |
| insect intestinal mucin 2 | 6 | 32 | 7.3 | 121.82 | XM_021326099.1 |
| Active hydrolases | |||||
| chymotrypsin-like protease C9 | 5 | 8 | 21.1 | 38.71 | XM_021335717.1 |
| trypsin, alkaline C-like | 1 | 1 | 3.9 | 27.167 | XM_021337856.1 |
| trypsin | 1 | 4 | 4.3 | 27.749 | XM_021337869.1 |
| trypsin-like protease | 2 | 2 | 11.9 | 26.816 | XM_021337877.1 |
| trypsin-like protease | 2 | 3 | 7.5 | 26.86 | XM_021337879.1 |
| trypsin | 1 | 1 | 6.5 | 26.217 | XM_021338513.1 |
| trypsin-7-like | 1 | 0 | 2.4 | 32.302 | XM_021340310.1 |
| trypsin T2a | 3 | 8 | 14.3 | 27.81 | XM_021340589.1 |
| trypsin | 1 | 4 | 2.7 | 27.549 | XM_021340592.1 |
| trypsin-like protease | 5 | 13 | 23.9 | 27.503 | XM_021340596.1 |
| trypsin-like protease | 7 | 142 | 44.9 | 26.916 | XM_021340599.1 |
| trypsin T4 | 2 | 6 | 11 | 26.772 | XM_021340602.1 |
| trypsin 2 | 1 | 2 | 3.3 | 32.037 | XM_021344969.1 |
| chymotrypsin | 3 | 3 | 14.6 | 30.518 | XM_021345778.1 |
| chymotrypsinogen | 6 | 2 | 21.4 | 30.834 | XM_021345781.1 |
| chymotrypsin-like protease C8 | 1 | 4 | 6.2 | 30.106 | XM_021345791.1 |
| chymotrypsin-like protease | 2 | 6 | 13.7 | 32.376 | XM_021345818.1 |
| chymotrypsin-like protease | 1 | 2 | 8.6 | 29.92 | XM_021345819.1 |
| carboxypeptidase | |||||
| carboxypeptidase B-like | 2 | 3 | 4.4 | 48.983 | XM_021328067.1 |
| carboxypeptidase precursor | 1 | 1 | 1.9 | 47.903 | XM_021330765.1 |
| carboxypeptidase B precursor | 2 | 3 | 5.1 | 48.318 | XM_021330831.1 |
| carboxypeptidase A | 1 | 2 | 2.8 | 48.526 | XM_021330834.1 |
| carboxypeptidase | 1 | 1 | 1.6 | 42.284 | XM_021330844.1 |
| carboxypeptidase | 3 | 5 | 10.3 | 47.935 | XM_021330848.1 |
| aminopeptidase N | 5 | 14 | 5 | 114.37 | XM_021337080.1 |
| aminopeptidase N | 4 | 8 | 4.2 | 112.81 | XM_021337081.1 |
| alpha-amylase | 1 | 1 | 2 | 56.009 | XM_021332568.1 |
| Inactive hydrolases | |||||
| serine protease inhibitor 5 | 1 | 2 | 2.8 | 45.123 | XM_021327264.1 |
| serine protease 24 | 2 | 2 | 6.6 | 43.33 | XM_021330937.1 |
| serine protease inhibitor | 1 | 1 | 1.6 | 88.323 | XM_021334028.1 |
| diverged serine protease | 4 | 13 | 13.3 | 27.289 | XM_021338518.1 |
| transmembrane protease serine 9-like | 2 | 18 | 2.5 | 79.868 | XM_021338520.1 |
| serine protease | 6 | 37 | 45.7 | 26.952 | XM_021340600.1 |
| serine protease inhibitor 3 isoform X1 | 1 | 1 | 1.5 | 106.56 | XM_021343251.1 |
| diverged serine protease | 5 | 13 | 22.1 | 25.554 | XM_021344422.1 |
| serine protease 52 | 1 | 3 | 4.7 | 27.001 | XM_021344467.1 |
| serine protease 3, partial | 3 | 5 | 10.7 | 30.739 | XM_021345780.1 |
| lipase | 3 | 9 | 14.2 | 30.739 | XM_021326676.1 |
| neutral lipase | 1 | 1 | 3.3 | 36.909 | XM_021331172.1 |
| neutral lipase | 1 | 2 | 3.6 | 36.476 | XM_021331174.1 |
| neutral lipase | 2 | 5 | 7.2 | 36.476 | XM_021331175.1 |
| pancreatic lipase 2 | 2 | 2 | 5.7 | 38.096 | XM_021338421.1 |
| lipase | 1 | 1 | 2.4 | 36.224 | XM_021338439.1 |
| pancreatic lipase 2 | 2 | 1 | 8.7 | 36.224 | XM_021343929.1 |
| neutral lipase | 1 | 2 | 3.6 | 35.29 | XM_021344621.1 |
| inactive lipase | 1 | 2 | 5.7 | 30.895 | XM_021344707.1 |
| Immune-related | |||||
| tubulin alpha-1 chain-like | 1 | 2 | 2 | 49.819 | XM_021338879.1 |
| Lipocalins | |||||
| fatty acid-binding protein 3 | 4 | 4 | 33.3 | 14.744 | XM_021333801.1 |
| fatty acid-binding protein 2 | 2 | 2 | 16.4 | 15.066 | XM_021341051.1 |
| fatty acid-binding protein 1 | 1 | 1 | 9.7 | 14.986 | XM_021341061.1 |
| fatty acid-binding protein 2 | 2 | 3 | 4 | 101.47 | XM_021341064.1 |
| polycalin | 1 | 2 | 0.8 | 101.47 | XM_021334936.1 |
| Hexamerins | |||||
| arylphorin | 8 | 19 | 11 | 82.226 | XM_021340131.1 |
| arylphorin | 5 | 4 | 4.9 | 82.28 | XM_021340132.1 |
| heat shock protein | |||||
| heat shock protein 90 | 2 | 2 | 1.8 | 82.63 | XM_021341131.1 |
| heat shock protein | 3 | 7 | 5.5 | 73.029 | XM_021332476.1 |
| others |
| Gene ID | FC a | Padj Value | mRNA FC | Description |
|---|---|---|---|---|
| XM_021332568.1 | 21.08226 | 0.033424 | 0.24 | alpha-amylase 2-like |
| XM_021341184.1 | 2.195151 | 0.03154 | 0.13 | chitin deacetylase 5α |
| XM_021328621.1 | 1.576713 | 0.0048 | −0.45 | unconventional myosin-XV isoform X2 |
| XM_021337877.1 | −1.86068 | 0.006606 | −0.91 | trypsin-like protease |
| XM_021337869.1 | −2.20431 | 0.018165 | −7.22 | trypsin |
| XM_021340592.1 | −2.73246 | 0.025215 | −1.36 | trypsin |
| XM_021337879.1 | −4.27566 | 0.035587 | - | trypsin-like protease |
| XM_021330834.1 | −22.6429 | 0.000219 | 1.40 | carboxypeptidase A |
| XM_021340037.1 | −23.7458 | 0.011483 | −1.53 | uncharacterized protein |
| XM_021345819.1 | −24.6671 | 0.036741 | −0.62 | chymotrypsin-like protease |
| XM_021345818.1 | −25.2913 | 0.032925 | −0.02 | chymotrypsin-like protease |
| XM_021340602.1 | −28.7418 | 0.017556 | −3.56 | trypsin T4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, M.; Liao, C.; Chakrabarty, S.; Wu, K.; Xiao, Y. Comparative Proteomics of Peritrophic Matrix Provides an Insight into its Role in Cry1Ac Resistance of Cotton Bollworm Helicoverpa armigera. Toxins 2019, 11, 92. https://doi.org/10.3390/toxins11020092
Jin M, Liao C, Chakrabarty S, Wu K, Xiao Y. Comparative Proteomics of Peritrophic Matrix Provides an Insight into its Role in Cry1Ac Resistance of Cotton Bollworm Helicoverpa armigera. Toxins. 2019; 11(2):92. https://doi.org/10.3390/toxins11020092
Chicago/Turabian StyleJin, Minghui, Chongyu Liao, Swapan Chakrabarty, Kongming Wu, and Yutao Xiao. 2019. "Comparative Proteomics of Peritrophic Matrix Provides an Insight into its Role in Cry1Ac Resistance of Cotton Bollworm Helicoverpa armigera" Toxins 11, no. 2: 92. https://doi.org/10.3390/toxins11020092
APA StyleJin, M., Liao, C., Chakrabarty, S., Wu, K., & Xiao, Y. (2019). Comparative Proteomics of Peritrophic Matrix Provides an Insight into its Role in Cry1Ac Resistance of Cotton Bollworm Helicoverpa armigera. Toxins, 11(2), 92. https://doi.org/10.3390/toxins11020092