An Integrative Analysis of Transcriptomics and Proteomics Reveals Novel Insights into the Response in the Midgut of Spodoptera frugiperda Larvae to Vip3Aa
Abstract
:1. Introduction
2. Results
2.1. Vip3Aa Significantly Inhibited the Growth of S. frugiperda
2.2. Transcriptomic Analysis of Midguts from Different Treatments
2.3. Gene Set Enrichment Analysis (GSEA)
2.4. Trend Analysis
2.5. Proteomic Profiling of S. frugiperda’s Response to Vip3Aa
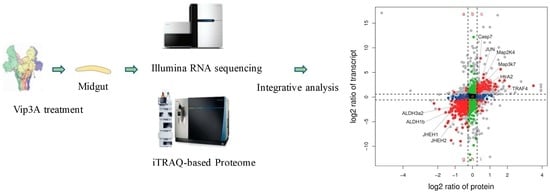
2.6. Correlation Analysis the Transcriptomic and Proteomic Data
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. Treatment by Vip3Aa
4.3. RNA Extraction and cDNA Library Construction
4.4. The Analysis of RNA-seq Data
4.5. Gene Set Enrichment Analysis (GSEA) and Trend Analysis
4.6. iTRAQ-Based Proteome Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jurat-Fuentes, J.L.; Heckel, D.G.; Ferre, J. Mechanisms of resistance to insecticidal proteins from Bacillus thuringiensis. Annu. Rev. Entomol. 2021, 66, 121–140. [Google Scholar] [CrossRef]
- Shelton, A.M.; Zhao, J.Z.; Roush, R.T. Economic, ecological, food safety, and social consequences of the deployment of Bt transgenic plants. Annu. Rev. Entomol. 2002, 47, 845–881. [Google Scholar] [CrossRef]
- Heckel, D.G. How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective. Arch. Insect Biochem. Physiol. 2020, 104, e21673. [Google Scholar] [CrossRef] [Green Version]
- Estruch, J.J.; Warren, G.W.; Mullins, M.A.; Nye, G.J.; Craig, J.A.; Koziel, M.G. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. USA 1996, 93, 5389–5394. [Google Scholar] [CrossRef] [Green Version]
- Chakroun, M.; Banyuls, N.; Bel, Y.; Escriche, B.; Ferre, J. Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 329–350. [Google Scholar] [CrossRef] [Green Version]
- Tabashnik, B.E.; Carriere, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.B.; Lu, G.Q.; Cheng, H.M.; Liu, C.X.; Xiao, Y.T.; Xu, C.; Shen, Z.C.; Wu, K.M. Transgenic cotton coexpressing Vip3A and Cry1Ac has a broad insecticidal spectrum against lepidopteran pests. J. Invertebr. Pathol. 2017, 149, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Walters, F.S.; Hart, H.; Palekar, N.; Chen, J.S. The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3A differs from that of Cry1Ab δ-endotoxin. Appl. Environ. Microbiol. 2003, 69, 4648–4657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, G.; Sachdev, B.; Sharma, N.; Seth, R.; Bhatnagar, R.K. Interaction of Bacillus thuringiensis vegetative insecticidal protein with ribosomal S2 protein triggers larvicidal activity in Spodoptera frugiperda. Appl. Environ. Microb. 2010, 76, 7202–7209. [Google Scholar] [CrossRef] [Green Version]
- Jiang, K.; Hou, X.Y.; Tan, T.T.; Cao, Z.L.; Mei, S.Q.; Yan, B.; Chang, J.; Han, L.; Zhao, D.; Cai, J. Scavenger receptor-C acts as a receptor for Bacillus thuringiensis vegetative insecticidal protein Vip3Aa and mediates the internalization of Vip3Aa via endocytosis. PLoS Pathog. 2018, 14, e1007347. [Google Scholar] [CrossRef]
- Jiang, K.; Hou, X.Y.; Han, L.; Tan, T.T.; Cao, Z.L.; Cai, J. Fibroblast growth factor receptor, a novel receptor for vegetative insecticidal protein Vip3Aa. Toxins 2018, 10, 546. [Google Scholar] [CrossRef] [Green Version]
- Jiang, K.; Mei, S.Q.; Wang, T.T.; Pan, J.H.; Chen, Y.H.; Cai, J. Vip3Aa induces apoptosis in cultured Spodoptera frugiperda (Sf9) cells. Toxicon 2016, 120, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Martinez, P.; Gomis-Cebolla, J.; Ferre, J.; Escriche, B. Changes in gene expression and apoptotic response in Spodoptera exigua larvae exposed to sublethal concentrations of Vip3 insecticidal proteins. Sci. Rep. 2017, 7, 16245. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.Y.; Han, L.; An, B.J.; Zhang, Y.L.; Cao, Z.L.; Zhan, Y.D.; Cai, X.; Yan, B.; Cai, J. Mitochondria and lysosomes participate in Vip3Aa-induced Spodoptera frugiperda Sf9 cell apoptosis. Toxins 2020, 12, 116. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.H.; Yang, Y.C.; Shan, Y.X.; Chakrabarty, S.; Cheng, Y.; Soberon, M.; Bravo, A.; Liu, K.Y.; Wu, K.M.; Xiao, Y.T. Two ABC transporters are differentially involved in the toxicity of two Bacillus thuringiensis Cry1 toxins to the invasive crop-pest Spodoptera frugiperda (J. E. Smith). Pest Manag. Sci. 2021, 77, 1492–1501. [Google Scholar] [CrossRef]
- Sun, X.X.; Hu, C.X.; Jia, H.R.; Wu, Q.L.; Shen, X.J.; Zhao, S.Y.; Jiang, Y.Y.; Wu, K.M. Case study on the first immigration of fall armyworm, Spodoptera frugiperda invading into China. J. Integr. Agric. 2021, 20, 664–672. [Google Scholar] [CrossRef]
- Bernardi, O.; Bernardi, D.; Horikoshi, R.J.; Okuma, D.M.; Miraldo, L.L.; Fatoretto, J.; Medeiros, F.C.L.; Burd, T.; Omoto, C. Selection and characterization of resistance to the Vip3Aa20 protein from Bacillus thuringiensis in Spodoptera frugiperda. Pest Manag. Sci. 2016, 72, 1794–1802. [Google Scholar] [CrossRef]
- Yang, F.; Morsello, S.; Head, G.P.; Sansone, C.; Huang, F.N.; Gilreath, R.T.; Kerns, D.L. F-2 screen, inheritance and cross-resistance of field-derived Vip3A resistance in Spodoptera frugiperda (Lepidoptera: Noctuidae) collected from Louisiana, USA. Pest Manag. Sci. 2018, 74, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Bel, Y.; Jakubowska, A.K.; Costa, J.; Herrero, S.; Escriche, B. Comprehensive analysis of gene expression profiles of the beet armyworm Spodoptera exigua larvae challenged with Bacillus thuringiensis Vip3Aa Toxin. PLoS ONE 2013, 8, e81927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.J.; Kang, S.; Sun, D.; Gong, L.J.; Zhou, J.L.; Qin, J.Y.; Guo, L.; Zhu, L.H.; Bai, Y.; Ye, F.; et al. MAPK-dependent hormonal signaling plasticity contributes to overcoming Bacillus thuringiensis toxin action in an insect host. Nat. Commun. 2020, 11, 3003. [Google Scholar] [CrossRef]
- Li, K.; Yang, M.; Yuen, P.M.P.; Chik, K.W.; Li, C.K.; Shing, M.M.K.; Lam, H.K.B.; Fok, T.F. Thrombospondin-1 induces apoptosis in primary leukemia and cell lines mediated by CD36 and Caspase-3. Int. J. Mol. Med. 2003, 12, 995–1001. [Google Scholar] [CrossRef]
- Turner, N.C.; Seckl, M.J. A Therapeutic Target for smoking-associated lung cancer. Sci. Transl. Med. 2010, 2, 62ps56. [Google Scholar] [CrossRef] [PubMed]
- Pardo, O.E.; Latigo, J.; Jeffery, R.E.; Nye, E.; Poulsom, R.; Spencer-Dene, B.; Lemoine, N.R.; Stamp, G.W.; Aboagye, E.O.; Seckl, M.J. The fibroblast growth factor receptor inhibitor PD173074 blocks small cell lung cancer growth in vitro and in vivo. Cancer Res. 2009, 69, 8645–8651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Los, F.C.O.; Kao, C.Y.; Smitham, J.; McDonald, K.L.; Ha, C.; Peixoto, C.A.; Aroian, R.V. RAB-5-and RAB-11-dependent vesicle-trafficking pathways are required for plasma membrane repair after attack by bacterial pore-forming toxin. Cell Host Microbe 2011, 9, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.H.; Tao, J.H.; Li, Q.; Cheng, Y.; Sun, X.X.; Wu, K.M.; Xiao, Y.T. Genome editing of the SfABCC2 gene confers resistance to Cry1F toxin from Bacillus thuringiensis in Spodoptera frugiperda. J. Integr. Agric. 2021, 20, 815–820. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, M.H.; Liao, C.Y.; Chakrabarty, S.; Wu, K.M.; Xiao, Y.T. Comparative proteomics of peritrophic matrix provides an insight into its role in Cry1Ac resistance of cotton bollworm Helicoverpa armigera. Toxins 2019, 11, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, M.; Shan, Y.; Peng, Y.; Wang, P.; Li, Q.; Yu, S.; Zhang, L.; Xiao, Y. An Integrative Analysis of Transcriptomics and Proteomics Reveals Novel Insights into the Response in the Midgut of Spodoptera frugiperda Larvae to Vip3Aa. Toxins 2022, 14, 55. https://doi.org/10.3390/toxins14010055
Jin M, Shan Y, Peng Y, Wang P, Li Q, Yu S, Zhang L, Xiao Y. An Integrative Analysis of Transcriptomics and Proteomics Reveals Novel Insights into the Response in the Midgut of Spodoptera frugiperda Larvae to Vip3Aa. Toxins. 2022; 14(1):55. https://doi.org/10.3390/toxins14010055
Chicago/Turabian StyleJin, Minghui, Yinxue Shan, Yan Peng, Ping Wang, Qi Li, Songmiao Yu, Lei Zhang, and Yutao Xiao. 2022. "An Integrative Analysis of Transcriptomics and Proteomics Reveals Novel Insights into the Response in the Midgut of Spodoptera frugiperda Larvae to Vip3Aa" Toxins 14, no. 1: 55. https://doi.org/10.3390/toxins14010055
APA StyleJin, M., Shan, Y., Peng, Y., Wang, P., Li, Q., Yu, S., Zhang, L., & Xiao, Y. (2022). An Integrative Analysis of Transcriptomics and Proteomics Reveals Novel Insights into the Response in the Midgut of Spodoptera frugiperda Larvae to Vip3Aa. Toxins, 14(1), 55. https://doi.org/10.3390/toxins14010055