Functional Characterization of New Polyketide Synthase Genes Involved in Ochratoxin A Biosynthesis in Aspergillus Ochraceus fc-1
Abstract
:1. Introduction
2. Results
2.1. Sequence and Phylogenetic Analysis of the Two AoOTApks Proteins


2.2. Identification and Knockout of AoOTApks in A. Ochraceus

| Primer Name | Sequences (5ʹ to 3′) |
|---|---|
| AoOTApks-1-up-F | TCAGTAGACCAGTGAGGCGA |
| AoOTApks-1-up-R | CAAAATAGGCATTGATGTGTTGACCTCCCGGAGTTCGGGTGGTGATAG |
| AoOTApks-1-down-F | CTCGTCCGAGGGCAAAGGAATAGAGTAGAATCGCCCTCTCTTATGGCG |
| AoOTApks-1-down-R | GGGCTTGCTCAAAACTCTGC |
| AoOTApks-1-knock-F | GCTGGAATCGCGACAGAGTA |
| AoOTApks-1-knock-R | ACAGCCAAGCGCCAATTAGA |
| AoOTApks-1-out-F | ATGAGATACAGGAGCAAGC |
| AoOTApks-1-out-R | CACCAAGCAGCAGATGAT |
| AoOTApks-2-up-F | AATCCGTTCAGCTCCCCAAG |
| AoOTApks-2-up-R | CAAAATAGGCATTGATGTGTTGACCTCCGAAGGCATCGCCGTCAAATC |
| AoOTApks-2-down-F | CTCGTCCGAGGGCAAAGGAATAGAGTAGTACAGGTGCTACTTGCGTGG |
| AoOTApks-2-down-R | AGAAACGGTGTCCATCGTCC |
| AoOTApks-2-knock-F | GACAGATGGGATAGACGCCG |
| AoOTApks-2-knock-R | CTGAAAACGGGCAATTGGGG |
| AoOTApks-2-out-F | CACTCGGTCCTGCGTTAA |
| AoOTApks-2-out-R | GTCGTTCACTTACCTTGCTT |
| hph-F a | GGAGGTCAACACATCAATGCCTATT |
| hph-R a | CTACTCTATTCCTTTGCCCT |
| Primer Name | Sequences (5′ to 3′) |
|---|---|
| AoOTApks-1-RT-F | CGCCTCATCATCAATCCTT |
| AoOTApks-1-RT-R | CAACTCGGTCAAGCAGAT |
| AoOTApks-2-RT-F | GGTGTGGCTGATGTAGTG |
| AoOTApks-2-RT-R | GTCTGTGAAGGTGTATGAATAG |
| GADPH-RT-F | GACTCACTATGCTGCCTAC |
| GADPH-RT-R | GAACCTTCTTGCCGTTGA |
2.3. Ochratoxin A Production and Expression Analysis of AoOTApks in A. Ochraceus Grown on Corn Medium

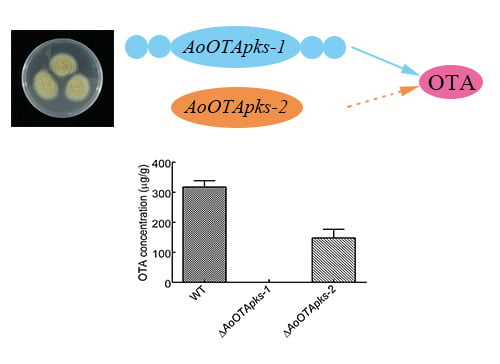
2.4. Production of OTA in Mutant and Wild Type Strains of A. Ochraceus



3. Discussion
4. Experimental Section
4.1. Fungal Strains and Culture Conditions
4.2. DNA and RNA Extraction, cDNA Synthesis, qRT-PCR
4.3. Amino Acid Alignments and Phylogenetic Analyses of OTA PKSs
4.4. Disruption of AoOTApks Genes and Construction of the ∆AoOTApks Mutants
4.5. Detection and Measurement of OTA by HPLC-FLD
4.6. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pfohl-Leszkowicz, A. Ochratoxin A and aristolochic acid involvement in nephropathies and associated urothelial tract tumours. Arh. Hig. Rada Toksikol. 2009, 60, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Petzinger, E.; Ziegler, K. Ochratoxin A from a toxicological perspective. J. Vet. Pharmacol. Ther. 2000, 23, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Denli, M.; Perez, J.F. Ochratoxins in feed, a risk for animal and human health: Control strategies. Toxins 2010, 2, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Pfohl-Leszkowicz, A.; Manderville, R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 61–99. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer (IARC), World Health Organization (WHO). Some naturally occurring substances: Food items and constituents, heterocyclic aromatic amines and mycotoxins. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 1993; Volume 56. [Google Scholar]
- Jørgensen, K. Occurrence of ochratoxin A in commodities and processed food—A review of EU occurrence data. Food Addit. Contam. 2005, 22 (Suppl. 1), 26–30. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, K. Survey of pork, poultry, coffee, beer and pulses for ochratoxin A. Food Addit. Contam. 2009, 15, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Farber, P.; Geisen, R. Analysis of differentially-expressed ochratoxin A biosynthesis genes of Penicillium nordicum. Eur. J. Plant Pathol. 2004, 110, 661–669. [Google Scholar] [CrossRef]
- Huff, W.; Hamilton, P. Mycotoxins-their biosynthesis in fungi: Ochratoxins-metabolites of combined pathways. J. Food Protect. 1979, 42, 815–820. [Google Scholar]
- Harris, J.P.; Mantle, P.G. Biosynthesis of ochratoxins by Aspergillus Ochraceus. Phytochemistry 2001, 58, 709–716. [Google Scholar] [CrossRef]
- Gallo, A.; Bruno, K.S.; Solfrizzo, M.; Perrone, G.; Mule, G.; Visconti, A.; Baker, S.E. New insight into the ochratoxin A biosynthetic pathway through deletion of a nonribosomal peptide synthetase gene in Aspergillus carbonarius. Appl. Environ. Microbiol. 2012, 78, 8208–8218. [Google Scholar] [CrossRef] [PubMed]
- Staunton, J. Polyketide biosynthesis: A millennium review. Natl. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef]
- Gallo, A.; Ferrara, M.; Perrone, G. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins 2013, 5, 717–742. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 6: Recent updates and new developments. Nucleic Acids Res. 2009, 37, D229–D232. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.; Yin, D.; Dawood, D.H.; Liu, X.; Chen, Y.; Ma, Z. Functional characterization of FgERG3 and FgERG5 associated with ergosterol biosynthesis, vegetative differentiation and virulence of Fusarium graminearum. Fungal Genet. Biol. 2014, 68, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 1–11. [Google Scholar]
- Yandell, M.; Ence, D. A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012, 13, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Knox, B.P.; Bruno, K.S.; Solfrizzo, M.; Baker, S.E.; Perrone, G.; Gallo, A.; Knox, B.P.; Bruno, K.S.; Solfrizzo, M.; et al. Identification and characterization of the polyketide synthase involved in ochratoxin A biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2014, 179, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Pel, H.J.; de Winde, J.H.; Archer, D.B.; Dyer, P.S.; Hofmann, G.; Schaap, P.J.; Turner, G.; de Vries, R.P.; Albang, R.; Albermann, K.; et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007, 25, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
- Bacha, N.; Mathieu, F.; Liboz, T.; Lebrihi, A.; Bacha, N.; Mathieu, F.; Liboz, T.; Lebrihi, A. Polyketide synthase gene aolc35–12 controls the differential expression of ochratoxin A gene aoks1 in Aspergillus westerdijkiae. World Mycotoxin J. 2012, 5, 177–186. [Google Scholar] [CrossRef]
- Geisen, R.; Mayer, Z.; Karolewiez, A.; Farber, P.F. Development of a real time PCR system for detection of Penicillium nordicum and for monitoring ochratoxin A production in foods by targeting the ochratoxin polyketide synthase gene. Syst. Appl. Microbiol. 2004, 27, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Geisen, R.; Schmidt-Heydt, M.; Karolewiez, A. A gene cluster of the ochratoxin A biosynthetic genes in Penicillium. Mycotoxin Res. 2006, 22, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Houbraken, J.A.M.P.; Samson, R.A.; Frank, J.M.; Kuijpers, A.F.A. New ochratoxin A producing species of Aspergillus section circumdati. Stud. Mycol. 2004, 18, 23–43. [Google Scholar]
- Gallo, A.; Perrone, G.; Solfrizzo, M.; Epifani, F.; Abbas, A.; Dobson, A.D.W.; Mule, G. Characterisation of a pks gene which is expressed during ochratoxin A production by Aspergillus carbonarius. Int. J. Food Microbiol. 2009, 129, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Gaffoor, I.; Trail, F. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl. Environ. Microbiol. 2006, 72, 1793–1799. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.T.; Lee, Y.R.; Jin, J.; Han, K.H.; Kim, H.; Kim, J.C.; Lee, T.; Yun, S.H.; Lee, Y.W. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol. Microbiol. 2005, 58, 1102–1113. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, L.; Davis, C.R.; Roach, C.; Nguyen, D.K.; Aldrich, T.; Mcada, P.C.; Reeves, C.D. Lovastatin biosynthesis in Aspergillus terreus: Characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem. Biol. 1999, 6, 429–439. [Google Scholar] [CrossRef]
- Kennedy, J.; Auclair, K.; Kendrew, S.G.; Park, C.; Vederas, J.C.; Richard Hutchinson, C. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 1999, 284, 1368–1372. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.E.; Kroken, S.; Inderbitzin, P.; Asvarak, T.; Li, B.-Y.; Shi, L.; Yoder, O.C.; Turgeon, B.G. Two polyketide synthase-encoding genes are required for biosynthesis of the polyketide virulence factor, T-toxin, by Cochliobolus heterostrophus. Mol. Plant-Microbe Interact. 2006, 19, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Suzuki, T.; Ono, C.; Iwamoto, K.; Hosobuchi, M.; Yoshikawa, H. Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol. Genet. Genomics 2002, 267, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.-M.; Szewczyk, E.; Davidson, A.D.; Keller, N.; Oakley, B.R.; Wang, C.C. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J. Am. Chem. Soc. 2009, 131, 2965–2970. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hadi, A.; Carter, D.; Magan, N. Temporal monitoring of the nor-1 (aflD) gene of Aspergillus flavus in relation to aflatoxin B1 production during storage of peanuts under different water activity levels. J. Appl. Microbiol. 2010, 109, 1914–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Heydt, M.; Richter, W.; Michulec, M.; Buttinger, G.; Geisen, R. Molecular and chemical monitoring of growth and ochratoxin A biosynthesis of P. verrucosum in wheat stored at different moisture conditions. Mycotoxin Res. 2007, 23, 138–146. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Wang, Y.; Wang, Q.; Liu, F.; Selvaraj, J.N.; Liu, L.; Xing, F.; Zhao, Y.; Zhou, L.; Liu, Y. Functional Characterization of New Polyketide Synthase Genes Involved in Ochratoxin A Biosynthesis in Aspergillus Ochraceus fc-1. Toxins 2015, 7, 2723-2738. https://doi.org/10.3390/toxins7082723
Wang L, Wang Y, Wang Q, Liu F, Selvaraj JN, Liu L, Xing F, Zhao Y, Zhou L, Liu Y. Functional Characterization of New Polyketide Synthase Genes Involved in Ochratoxin A Biosynthesis in Aspergillus Ochraceus fc-1. Toxins. 2015; 7(8):2723-2738. https://doi.org/10.3390/toxins7082723
Chicago/Turabian StyleWang, Liuqing, Yan Wang, Qi Wang, Fei Liu, Jonathan Nimal Selvaraj, Lingna Liu, Fuguo Xing, Yueju Zhao, Lu Zhou, and Yang Liu. 2015. "Functional Characterization of New Polyketide Synthase Genes Involved in Ochratoxin A Biosynthesis in Aspergillus Ochraceus fc-1" Toxins 7, no. 8: 2723-2738. https://doi.org/10.3390/toxins7082723
APA StyleWang, L., Wang, Y., Wang, Q., Liu, F., Selvaraj, J. N., Liu, L., Xing, F., Zhao, Y., Zhou, L., & Liu, Y. (2015). Functional Characterization of New Polyketide Synthase Genes Involved in Ochratoxin A Biosynthesis in Aspergillus Ochraceus fc-1. Toxins, 7(8), 2723-2738. https://doi.org/10.3390/toxins7082723